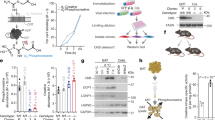
Abstract
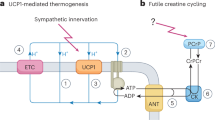
Adaptive thermogenesis has attracted much attention because of its ability to increase systemic energy expenditure and to counter obesity and diabetes1,2,3. Recent data have indicated that thermogenic fat cells use creatine to stimulate futile substrate cycling, dissipating chemical energy as heat4,5. This model was based on the super-stoichiometric relationship between the amount of creatine added to mitochondria and the quantity of oxygen consumed. Here we provide direct evidence for the molecular basis of this futile creatine cycling activity in mice. Thermogenic fat cells have robust phosphocreatine phosphatase activity, which is attributed to tissue-nonspecific alkaline phosphatase (TNAP). TNAP hydrolyses phosphocreatine to initiate a futile cycle of creatine dephosphorylation and phosphorylation. Unlike in other cells, TNAP in thermogenic fat cells is localized to the mitochondria, where futile creatine cycling occurs. TNAP expression is powerfully induced when mice are exposed to cold conditions, and its inhibition in isolated mitochondria leads to a loss of futile creatine cycling. In addition, genetic ablation of TNAP in adipocytes reduces whole-body energy expenditure and leads to rapid-onset obesity in mice, with no change in movement or feeding behaviour. These data illustrate the critical role of TNAP as a phosphocreatine phosphatase in the futile creatine cycle.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (https://www.ebi.ac.uk/pride/) with the dataset identifier PXD025032. The published adipocyte-specific ribosomal profiling dataset can be downloaded at https://ars.els-cdn.com/content/image/1-s2.0-S1550413118301839-mmc2.xlsx. Full scans for all western blots are provided in the Supplementary Information. All other data are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Rosen, E. D. & Spiegelman, B. M. What we talk about when we talk about fat. Cell 156, 20–44 (2014).
Harms, M. & Seale, P. Brown and beige fat: development, function and therapeutic potential. Nat. Med. 19, 1252–1263 (2013).
Carobbio, S., Guénantin, A. C., Samuelson, I., Bahri, M. & Vidal-Puig, A. Brown and beige fat: from molecules to physiology and pathophysiology. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1864, 37–50 (2019).
Chouchani, E. T., Kazak, L. & Spiegelman, B. M. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab. 29, 27–37 (2019).
Kazak, L. et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 163, 643–655 (2015).
Cypess, A. M. et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 19, 635–639 (2013).
Cypess, A. M. et al. Activation of human brown adipose tissue by a β3-adrenergic receptor agonist. Cell Metab. 21, 33–38 (2015).
Leitner, B. P. et al. Mapping of human brown adipose tissue in lean and obese young men. Proc. Natl Acad. Sci. USA 114, 8649–8654 (2017).
Bachman, E. S. et al. βAR signaling required for diet-induced thermogenesis and obesity resistance. Science 297, 843–845 (2002).
Zeng, X. et al. Innervation of thermogenic adipose tissue via a calsyntenin 3β–S100b axis. Nature 569, 229–235 (2019).
Enerbäck, S. et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 (1997).
Hofmann, W. E., Liu, X., Bearden, C. M., Harper, M. E. & Kozak, L. P. Effects of genetic background on thermoregulation and fatty acid-induced uncoupling of mitochondria in UCP1-deficient mice. J. Biol. Chem. 276, 12460–12465 (2001).
Kazak, L. et al. UCP1 deficiency causes brown fat respiratory chain depletion and sensitizes mitochondria to calcium overload-induced dysfunction. Proc. Natl Acad. Sci. USA 114, 7981–7986 (2017).
Ikeda, K. et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat. Med. 23, 1454–1465 (2017).
Long, J. Z. et al. The secreted enzyme PM20D1 regulates lipidated amino acid uncouplers of mitochondria. Cell 166, 424–435 (2016).
Kazak, L. et al. Genetic depletion of adipocyte creatine metabolism inhibits diet-induced thermogenesis and drives obesity. Cell Metab. 26, 660–671.e3 (2017).
Kazak, L. et al. Ablation of adipocyte creatine transport impairs thermogenesis and causes diet-induced obesity. Nat. Metab. 1, 360–370 (2019).
Rahbani, J. F. et al. Creatine kinase B controls futile creatine cycling in thermogenic fat. Nature 590, 480–485 (2021).
Müller, S. et al. Proteomic analysis of human brown adipose tissue reveals utilization of coupled and uncoupled energy expenditure pathways. Sci. Rep. 6, 30030 (2016).
Svensson, P. A. et al. Gene expression in human brown adipose tissue. Int. J. Mol. Med. 27, 227–232 (2011).
Bessman, S. P. & Carpenter, C. L. The creatine–creatine phosphate energy shuttle. Annu. Rev. Biochem. 54, 831–862 (1985).
Pinkerton, A. B. et al. Discovery of 5-((5-chloro-2-methoxyphenyl)sulfonamido)nicotinamide (SBI-425), a potent and orally bioavailable tissue-nonspecific alkaline phosphatase (TNAP) inhibitor. Bioorg. Med. Chem. Lett. 28, 31–34 (2018).
Roh, H. C. et al. Warming induces significant reprogramming of beige, but not brown, adipocyte cellular identity. Cell Metab. 27, 1121–1137.e5 (2018).
Buchet, R., Millán, J. L. & Magne, D. Multisystemic functions of alkaline phosphatases. Methods Mol. Biol. 1053, 27–51 (2013).
Hessle, L. et al. Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc. Natl Acad. Sci. USA 99, 9445–9449 (2002).
Anderson, H. C. et al. Impaired calcification around matrix vesicles of growth plate and bone in alkaline phosphatase-deficient mice. Am. J. Pathol. 164, 841–847 (2004).
Lam, S. S. et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods 12, 51–54 (2015).
Rhee, H. W. et al. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 339, 1328–1331 (2013).
Kiffer-Moreira, T. et al. Catalytic signature of a heat-stable, chimeric human alkaline phosphatase with therapeutic potential. PLoS ONE 9, e89374 (2014).
Chen, W. W., Freinkman, E., Wang, T., Birsoy, K. & Sabatini, D. M. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e11 (2016).
Chen, W. W., Freinkman, E. & Sabatini, D. M. Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat. Protoc. 12, 2215–2231 (2017).
Haarhaus, M., Brandenburg, V., Kalantar-Zadeh, K., Stenvinkel, P. & Magnusson, P. Alkaline phosphatase: a novel treatment target for cardiovascular disease in CKD. Nat. Rev. Nephrol. 13, 429–442 (2017).
Liu, X. et al. Paradoxical resistance to diet-induced obesity in UCP1-deficient mice. J. Clin. Invest. 111, 399–407 (2003).
Feldmann, H. M., Golozoubova, V., Cannon, B. & Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 9, 203–209 (2009).
Vendelin, M., Lemba, M. & Saks, V. A. Analysis of functional coupling: mitochondrial creatine kinase and adenine nucleotide translocase. Biophys. J. 87, 696–713 (2004).
Shulman, G. I., Ladenson, P. W., Wolfe, M. H., Ridgway, E. C. & Wolfe, R. R. Substrate cycling between gluconeogenesis and glycolysis in euthyroid, hypothyroid, and hyperthyroid man. J. Clin. Invest. 76, 757–764 (1985).
Wolfe, R. R., Herndon, D. N., Jahoor, F., Miyoshi, H. & Wolfe, M. Effect of severe burn injury on substrate cycling by glucose and fatty acids. N. Engl. J. Med. 317, 403–408 (1987).
Clark, M. G. et al. Accelerated substrate cycling of fructose-6-phosphate in the muscle of malignant hyperthermic pigs. Nature 245, 99–101 (1973).
Newsholme, E. A. & Crabtree, B. Substrate cycles in metabolic regulation and in heat generation. Biochem. Soc. Symp. 61–109 (1976).
Klein, S. & Wolfe, R. R. Whole-body lipolysis and triglyceride-fatty acid cycling in cachectic patients with esophageal cancer. J. Clin. Invest. 86, 1403–1408 (1990).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Klein, J. et al. β3-adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in brown adipocytes. J. Biol. Chem. 274, 34795–34802 (1999).
Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012).
Sharabi, K. et al. Selective chemical inhibition of PGC-1α gluconeogenic activity ameliorates type 2 diabetes. Cell 169, 148–160.e15 (2017).
Springer, M. L., Rando, T. A. & Blau, H. M. Gene delivery to muscle. Curr. Protoc. Hum. Genet. 31, 13.4.1–13.4.19 (2002).
Allan, C. et al. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245–253 (2012).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Yuan, M., Breitkopf, S. B., Yang, X. & Asara, J. M. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat. Protoc. 7, 872–881 (2012).
Foster, B. L. et al. Conditional Alpl ablation phenocopies dental defects of hypophosphatasia. J. Dent. Res. 96, 81–91 (2017).
Gettins, P., Metzler, M. & Coleman, J. E. Alkaline phosphatase. 31P NMR probes of the mechanism. J. Biol. Chem. 260, 2875–2883 (1985).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Mootha, V. K. et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003).
Acknowledgements
We thank C. J. Rosen for sharing the Alplfl/fl mouse strain with permission of J.L.M.; R. Garrity for help with CLAMS studies; the NMR Core jointly operated by Harvard Medical School and Dana-Farber Cancer Institute for help with NMR data acquisition; Nikon Imaging Center at Harvard Medical School for help with fluorescence imaging studies; the EM Core at Harvard Medical School for APEX2/EM imaging studies; and the Mass Spectrometry Facility at Beth Israel Deaconess Medical Center for targeted metabolomics studies. Y.S. and C.L.R. are supported by the American Heart Association postdoctoral fellowship. J.F.R. is supported by the Charlotte and Leo Karassik Fellowship. B.H. was a Cancer Research Institute/Leonard Kahn Foundation Fellow. P.A.D. is supported by a Damon Runyon Cancer Research Foundation Fellowship. This study was supported by NIDCR grant DE12889 to J.L.M., NIH grant DK 123095 to E.T.C., Canadian Institutes of Health Research (CIHR) grant PJT-159529 to L.K. and JPB Foundation 6293803 and NIH grant DK123228 to B.M.S.
Author information
Authors and Affiliations
Contributions
Y.S. and B.M.S. conceived the study and designed the experiments. Y.S. performed experiments and analysed the data. Y.S. and J.F.R. performed cellular and mitochondrial respiration experiments and analysed the data. M.P.J. performed mass spectrometry analysis. C.L.R. and S.V. helped with fluorescence imaging studies. S.V. assisted with protease protection assays. D.B. assisted with all experiments. B.H. assisted with animal experiments. C.L.R., S.V., P.A.D., X.Z., A.B.W. and N.H.K. helped with animal and/or cellular experiments and data analyses. C.R.K. assisted with CLAMS studies. A.M. assisted with activity measurements and animal experiments. J.L.M. contributed essential genetic and pharmacological reagents and discussed data. L.K. and E.T.C. contributed to experimental discussions. Y.S. and B.M.S. wrote the manuscript with comments from all authors. All authors provided input and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 PCr phosphatase activities of thermogenic fat and TNAP.
a, PCr phosphatase activities of total mitochondrial protein extracts from different tissues of cold-acclimated mice. Mitochondrial protein extract was prepared from tissues excised from 10 mice for BAT or 20 mice for iWAT. Each reaction contains 10 mM of PCr and 0.4 mg ml−1 of mitochondrial protein extract, except the buffer control. Data are presented as the estimated parameters ± uncertainties. Uncertainties are represented by the standard errors of nonlinear regression that fits a straight-line model to the initial linear phase of PCr hydrolysis kinetics measured by 31P NMR over 11 time points for BAT and iWAT and 6 time points for the buffer control (shown in Source Data). b, Ion-exchange chromatography of the active fraction of SEC. The PCr phosphatase activity of each fraction was measured by enzyme-coupled assay. The activity of the most active fraction was also verified by 31P NMR. Red and blue bars denote the fractions used for isobaric labelling (TMT) and quantitative mass spectrometric analysis. c, Western-blot analysis of the active SEC fraction prepared from cold-acclimated mice (cold), compared with the equivalent fraction prepared from room-temperature housed mice (RT). d, PCr phosphatase activities of total mitochondrial protein extracts from BAT of cold-acclimated mice treated with vehicle or SBI-425 (10 μM), measured by 31P NMR. n = 2 technical replicates per group. Data are presented as mean ± s.e.m. e, Stacked traces of 31P NMR spectra recorded at indicated time points, demonstrating the kinetics of PCr hydrolysis catalysed by recombinant TNAP. The minor peak marked with an asterisk on top is from glycerol-3-phosphate, a side-product of the phospho-transferase activity of TNAP50, that transfers the phosphoryl-group from PCr to glycerol present in the reaction buffer. f, Michaelis constant (Km) curves of hydrolysis of PCr (left) and PPi (right) catalysed by recombinant TNAP. Activities were measured by the enzyme-coupled assay; n = 2 technical replicates. Data are presented as mean ± s.e.m. g, Comparison of the Michaelis–Menten parameters extrapolated from f. Data are presented as the estimated parameters ± uncertainties. Uncertainties are represented by standard errors derived from the nonlinear regression fit of Michaelis–Menten model to the data in f.
Extended Data Fig. 2 Mitochondrial localization of ectopically expressed TNAP in non-thermogenic fat cell types.
Confocal fluorescence microscopic images showing subcellular localization of ectopically expressed TNAP in different cell types. PTEC, kidney proximal tubule epithelial cells. The insets show a magnified view of the area outlined by the dotted box. Anti-TNAP and anti-HSP60 were used to visualize TNAP and mitochondria, respectively. Scale bars, 10 μm.
Extended Data Fig. 3 Mitochondrial localization of endogenous TNAP in BAT and non-thermogenic fat cells.
a, Confocal fluorescence images showing subcellular localization of endogenous TNAP in brown adipocytes (top and middle panels) and hepatocytes (bottom panels). Primary brown preadipocytes were prepared from Alplfl/fl mice, transduced with either AdGFP (WT) or AdCre (Alpl KO) on day 4 of differentiation, and fixed for imaging on day 8. Arrows denote selected peri-nuclear areas of TNAP signal that co-localize with mitochondria signal. Antibodies for TNAP (red) and HSP60 (green) were used to visualize TNAP and mitochondria. Scale bar, 5 μm. b, Pearson’s correlation coefficient (PCC) analysis showing the extent of co-localization of TNAP with mitochondria in indicated cell types; n = 10 cells per group; data are presented as mean ± s.e.m.; statistical significance was calculated by one-way ANOVA with Bonferroni’s multiple comparisons test. c, Western-blot analysis on TNAP in wild-type versus knockout cells. Vinculin (VCL) blot was used as a sample preparation control. d, Confocal fluorescence microscopic images showing subcellular localization of endogenous TNAP in different cell types. Anti-TNAP and anti-HSP60 were used to visualize TNAP and mitochondria, respectively. Scale bars, 5 μm. e, Western-blot analysis on TNAP and mitochondrial markers in mitochondrial preparations from BAT of cold-acclimated, wild-type vs adipo-Alpl knockout mice. Blots were processed in parallel with samples derived from the same experiment. f, Western-blot analysis of the insoluble fraction of mitochondria extract treated with phosphatidylinositol-specific phospholipase-C (PI-PLC), followed by ultracentrifugation, showing that PLC treatment releases TNAP from membranes. P, pellet; S, supernatant. Mitochondrial preparation was fragmented by sonication before treatment. Blots were processed in parallel with samples derived from the same experiment.
Extended Data Fig. 4 Proximity-based fluorescent labelling by TNAP–APEX2 and trypsin protection assay on mitochondria from BAT.
a, Confocal fluorescence microscopic images of immortalized brown adipocytes showing co-localization of the GFP signal from 3XHA-EGFP-OMP25 construct (mGFP, channel: 488 nm) with different mitochondria markers. Endogenous antibodies, OxPhos (upper red, channel: 561 nm) and HSP60 (lower red, channel: 640 nm), were used to visualize mitochondria. The insets show a magnified region of the image outlined by the dotted box. Scale bars, 5 μm. b, Illustration of how APEX2 reports subcellular localization of TNAP by its peroxidase activity. X indicates either Alexa Fluor 647-conjugated tyramide (for confocal microscopy) or 3,3′-diaminobenzidine (for TEM studies). c, Confocal fluorescence analysis of immortalized brown adipocytes (top) and hepatocytes (bottom) ectopically expressing a TNAP–APEX2 construct. Cells were fixed and treated with Alexa Fluor 647–tyramide/H2O2 for proximity-based fluorescent labelling facilitated by the peroxidase activity of APEX2. Stably expressed 3XHA-EGFP-OMP25 was used as mitochondria reporter. Scale bars, 10 μm. d, Western blot analysis of the protease protection assay on mitochondria derived from BAT of cold-acclimated mice. TOMM20, GPD2 (glycerol-3-phosphate dehydrogenase), Cyt C (cytochrome c) and CS (citrate synthase) are shown as markers of outer mitochondrial membrane (OMM), intermembrane space (IMS) and mitochondrial matrix. Blots were processed in parallel with samples derived from the same experiment. e, Relative protein abundances in trypsin-digested mitochondria derived from band intensities of intact protein quantified from d; n = 2 technical replicates. Data are presented as mean ± s.e.m.
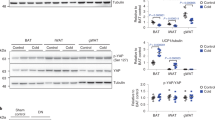
Extended Data Fig. 5 Effect of Alpl silencing on cellular respiration.
a, Quantitative PCR with reverse transcription (qRT–PCR) of differentiated primary brown preadipocytes treated with shLacZ or shAlpl; n = 3 biologically independent samples per group. b, Western-blot analysis on TNAP in cells treated with shLacZ or shAlpl. Vinculin (VCL) blot was used as a sample processing control. c, Effect of Alpl knockdown (adenoviral shAlpl) on the oxygen consumption rate (OCR) of primary brown adipocytes. Treatments to initiate different respiration states are as follows: stimulated, noradrenaline; uncoupled, oligomycin; maximum, carbonyl cyanide m-chlorophenyl hydrazone; n = 11 biologically independent samples for the shLacZ group and 18 biologically independent samples for the shAlpl group. Data are presented as mean ± s.e.m. Statistical significance was calculated by unpaired Student’s two-sided t-test.
Extended Data Fig. 6 Effect of TNAP inhibition on the futile creatine cycle.
a, Effect of SBI-425 treatment (10 μM) on the OCR of beige fat-derived mitochondria from wild-type vs adipo-Alpl knockout mice in the presence of 0.01 mM creatine and 0.1 mM ADP (limiting ADP) or 1 mM ADP (saturating ADP), as measured by a Seahorse XF24 Extracellular Flux Analyzer; n = 7 independent measurements per group. b, Effect of SBI-425 treatment (10 μM) on OCR of beige fat-derived mitochondria in the presence of 0.1 mM ADP (limiting ADP) or 1 mM ADP (saturating ADP), but in the absence of creatine, as measured by a Seahorse XF24 Extracellular Flux Analyzer; n = 6 independent measurements per group. Data are presented as mean ± s.e.m. Statistical significance was calculated by either two-way ANOVA with Bonferroni’s multiple comparisons test (a) or unpaired Student’s two-sided t-test (b).
Extended Data Fig. 7 Rapid mitochondria purification enriched mitochondrial metabolites and proteins.
a, Relative abundances of citric acid cycle intermediates in mitochondria vs whole-cell metabolomics; n = 6 biologically independent samples. Data are presented as mean ± s.e.m. b, Western-blot analysis on mitochondrial markers in immunoprecipitated mitochondria for metabolomics study. Blots were processed in parallel with samples derived from the same experiment.
Extended Data Fig. 8 Movement and food intake of mice upon SBI-425 treatment.
a, b, Cumulative movement and food intake of wild-type mice (a) and adipo-Alpl knockout mice (b) for 24 h after treatment of SBI-425 versus vehicle; n = 10 mice for wild-type and 4 mice for adipo-Alpl knockout. Data are presented as mean ± s.e.m. Statistical significance was calculated using an unpaired Student’s two-sided t-test.
Extended Data Fig. 9 Energy expenditure and movement of wild-type and adipo-Alpl knockout mice on a HFD.
a, Indirect calorimetric measurements taken for wild-type and adipo-Alpl knockout mice that had been on a HFD for 4 weeks at 22 °C; n = 7 mice for wild-type and 6 mice for adipo-Alpl knockout; the grey area indicates the dark period. b. Averaged respiration rates over 24 h as measured in a; n = 7 mice for wild-type and 6 mice for adipo-Alpl knockout. c, Cumulative movement of mice over 24 h; n = 7 mice for wild-type and 6 mice for adipo-Alpl knockout. Data are presented as mean ± s.e.m. Statistical significance was calculated by either two-way ANOVA (a) or unpaired Student’s two-sided t-test (b, c).
Extended Data Fig. 10 Compensatory thermogenesis in adipo-Alpl knockout mice.
a, Indirect calorimetric measurement taken for wild-type and adipo-Alpl knockout mice kept in metabolic cages at 22 °C, showing stimulation of respiration by CL 316,243 administration (1.0 mg kg−1). Arrow denotes the time point of drug administration; n = 12 mice for wild-type and 9 mice for adipo-Alpl knockout; all mice were pre-treated with CL 316,243 (1.0 mg kg−1 day−1) for 5 days. b, Western-blot analysis of BAT and iWAT from wild-type or adipo-Alpl knockout mice pre-treated with CL 316,243 (1.0 mg kg−1 day−1) for 5 days. c, d, Gene set enrichment plot of quantitative mass spectrometric analyses of BAT (c) and iWAT (d) from wild-type or adipo-Alpl knockout mice treated with CL 316,243 (1.0 mg kg−1 day−1) for 5 days; n = 4 mice for wild-type and 6 mice for adipo-Alpl knockout. Enrichment analysis was performed with GSEA 4.1.051,52. The hallmark gene sets were surveyed, and oxidative phosphorylation is the top hit for both BAT and iWAT. Family-wise error rate P value was presented for statistical significance (number of permutations, 1,000); NES, normalized enrichment score (enrichment statistics, ‘classic’). e, f, qRT–PCR of BAT (e) and iWAT (f) from wild-type and adipo-Alpl knockout mice treated with CL 316,243 (1.0 mg kg−1 day−1) for 5 days; n = 4 mice for wild-type and 6 mice for adipo-Alpl knockout. Data are presented as mean ± s.e.m. Statistical significance was calculated by either two-way ANOVA (a) or unpaired Student’s two-sided t-test (e, f).
Supplementary information
Supplementary Figure 1
This file contains the uncropped images of western blots represented in Extended Data Figure 1c, 3c, 3e-f, 4d, 5b, 7b, and 10b. Red boxes indicate regions displayed in the final figures.
Supplementary Table 1
This file contains the mitochondrial metabolomics data of brown adipocytes treated with vehicle vs SBI-425 (10 μM) for 1 hour.
Supplementary Table 2
This file contains the proteomics data of BAT and iWAT from WT vs Adipo-Alpl KO mice housed at room-temperature and pre-treated with CL 316,243 (1.0 mg/kg/day) for 5 days.
Source data
Rights and permissions
About this article
Cite this article
Sun, Y., Rahbani, J.F., Jedrychowski, M.P. et al. Mitochondrial TNAP controls thermogenesis by hydrolysis of phosphocreatine. Nature 593, 580–585 (2021). https://doi.org/10.1038/s41586-021-03533-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03533-z
This article is cited by
-
Futile lipid cycling: from biochemistry to physiology
Nature Metabolism (2024)
-
Is Propolis a Potential Anti-Obesogenic Agent for Obesity?
Current Nutrition Reports (2024)
-
A critical assessment of the role of creatine in brown adipose tissue thermogenesis
Nature Metabolism (2023)
-
The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase
Nature Communications (2023)
-
A general overview of the multifactorial adaptation to cold: biochemical mechanisms and strategies
Brazilian Journal of Microbiology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.