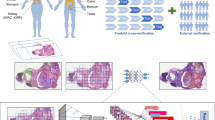
Abstract
Cancer of unknown primary (CUP) origin is an enigmatic group of diagnoses in which the primary anatomical site of tumour origin cannot be determined1,2. This poses a considerable challenge, as modern therapeutics are predominantly specific to the primary tumour3. Recent research has focused on using genomics and transcriptomics to identify the origin of a tumour4,5,6,7,8,9. However, genomic testing is not always performed and lacks clinical penetration in low-resource settings. Here, to overcome these challenges, we present a deep-learning-based algorithm—Tumour Origin Assessment via Deep Learning (TOAD)—that can provide a differential diagnosis for the origin of the primary tumour using routinely acquired histology slides. We used whole-slide images of tumours with known primary origins to train a model that simultaneously identifies the tumour as primary or metastatic and predicts its site of origin. On our held-out test set of tumours with known primary origins, the model achieved a top-1 accuracy of 0.83 and a top-3 accuracy of 0.96, whereas on our external test set it achieved top-1 and top-3 accuracies of 0.80 and 0.93, respectively. We further curated a dataset of 317 cases of CUP for which a differential diagnosis was assigned. Our model predictions resulted in concordance for 61% of cases and a top-3 agreement of 82%. TOAD can be used as an assistive tool to assign a differential diagnosis to complicated cases of metastatic tumours and CUPs and could be used in conjunction with or in lieu of ancillary tests and extensive diagnostic work-ups to reduce the occurrence of CUP.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The TCGA diagnostic whole-slide data and corresponding labels are available from NIH genomic data commons (https://portal.gdc.cancer.gov/). The CPTAC histology data and corresponding labels are available from the TCIA CPTAC Pathology Portal (https://cancerimagingarchive.net/datascope/cptac/). Processed data that are included in the figures presented in the paper are available as source data. Restrictions apply to the availability of the raw in-house and external data, which were used with institutional permission through IRB approval for the current study, and are thus not publicly available. Please email all requests for academic use of raw and processed data to the corresponding author (and also include M.Y.L. (mlu16@bwh.harvard.edu)). All requests will be evaluated based on institutional and departmental policies to determine whether the data requested is subject to intellectual property or patient privacy obligations. Data can only be shared for non-commercial academic purposes and will require a formal material transfer agreement. Source data are provided with this paper.
Code availability
All code was implemented in Python using PyTorch as the primary deep learning package. All code and scripts to reproduce the experiments of this paper are available at https://github.com/mahmoodlab/TOAD.
References
Rassy, E. & Pavlidis, N. Progress in refining the clinical management of cancer of unknown primary in the molecular era. Nat. Rev. Clin. Oncol. 17, 541–554 (2020).
Varadhachary, G. R. & Raber, M. N. Cancer of unknown primary site. N. Engl. J. Med. 371, 757–765 (2014).
Massard, C., Loriot, Y. & Fizazi, K. Carcinomas of an unknown primary origin—diagnosis and treatment. Nat. Rev. Clin. Oncol. 8, 701–710 (2011).
Jiao, W. et al. A deep learning system accurately classifies primary and metastatic cancers using passenger mutation patterns. Nat. Commun. 11, 728 (2020).
Penson, A. et al. Development of genome-derived tumor type prediction to inform clinical cancer care. JAMA Oncol. 6, 84–91 (2020).
Grewal, J. K. et al. Application of a neural network whole transcriptome-based pan-cancer method for diagnosis of primary and metastatic cancers. JAMA Netw. Open 2, e192597 (2019).
Zhao, Y. et al. CUP-AI-Dx: a tool for inferring cancer tissue of origin and molecular subtype using RNA gene-expression data and artificial intelligence. EBioMedicine 61, 103030 (2020).
Shen, Y. et al. TOD-CUP: a gene expression rank-based majority vote algorithm for tissue origin diagnosis of cancers of unknown primary. Brief. Bioinformatics 22, 2106–2118 (2020).
Kerr, S. E. et al. Multisite validation study to determine performance characteristics of a 92-gene molecular cancer classifier. Clin. Cancer Res. 18, 3952–3960 (2012).
Hayashi, H. et al. Site-specific and targeted therapy based on molecular profiling by next-generation sequencing for cancer of unknown primary site: a nonrandomized phase 2 clinical trial. JAMA Oncol. 6, 1931–1938 (2020).
Nass, D. et al. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 19, 375–383 (2009).
Estrella, J. S., Wu, T. T., Rashid, A. & Abraham, S. C. Mucosal colonization by metastatic carcinoma in the gastrointestinal tract: a potential mimic of primary neoplasia. Am. J. Surg. Pathol. 35, 563–572 (2011).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Liu, Y. et al. A deep learning system for differential diagnosis of skin diseases. Nat. Med. 26, 900–908 (2020).
Lu, M. Y. et al. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-020-00682-w (2021).
Campanella, G. et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 25, 1301–1309 (2019).
Chen, P. C. et al. An augmented reality microscope with real-time artificial intelligence integration for cancer diagnosis. Nat. Med. 25, 1453–1457 (2019).
Ouyang, D. et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580, 252–256 (2020).
Hollon, T. C. et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat. Med. 26, 52–58 (2020).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Kalra, S. et al. Pan-cancer diagnostic consensus through searching archival histopathology images using artificial intelligence. NPJ Digit. Med. 3, 31 (2020).
Coudray, N. et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 24, 1559–1567 (2018).
Kather, J. N. et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 25, 1054–1056 (2019).
Poplin, R. et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat. Biomed. Eng. 2, 158–164 (2018).
Ilse, M., Tomczak, J. M. & Welling, M. Attention-based deep multiple instance learning. In International Conference on Machine Learning 2132–2141 (2018).
Handorf, C. R. et al. A multicenter study directly comparing the diagnostic accuracy of gene expression profiling and immunohistochemistry for primary site identification in metastatic tumors. Am. J. Surg. Pathol. 37, 1067–1075 (2013).
McHugh, M. L. Interrater reliability: the kappa statistic. Biochem. Med. 22, 276–282 (2012).
Sheahan, K. et al. Metastatic adenocarcinoma of an unknown primary site. A comparison of the relative contributions of morphology, minimal essential clinical data and CEA immunostaining status. Am. J. Clin. Pathol. 99, 729–735 (1993).
Jurmeister, P. et al. Machine learning analysis of DNA methylation profiles distinguishes primary lung squamous cell carcinomas from head and neck metastases. Sci. Transl. Med. 11, eaaw8513 (2019).
Rassy, E. & Pavlidis, N. The currently declining incidence of cancer of unknown primary. Cancer Epidemiol. 61, 139–141 (2019).
He, K. et al. Deep residual learning for image recognition. In Proc. IEEE Conference on Computer Vision and Pattern Recognition 770–778 (IEEE, 2016).
Russakovsky, O. et al. Imagenet large scale visual recognition challenge. Int. J. Comput. Vis. 115, 211–252 (2015).
Youden, W. J. Index for rating diagnostic tests. Cancer 3, 32–35 (1950).
Yosinski, J., Clune, J. Bengio, Y. & Lipson, H. How transferable are features in deep neural networks? In Proc. 27th International Conference on Neural Information Processing Systems Vol. 2, 3320–3328 (2014).
Graham, S. et al. Hover-net: simultaneous segmentation and classification of nuclei in multi-tissue histology images. Med. Image Anal. 58, 101563 (2019).
Acknowledgements
We thank A. Bruce for scanning internal cohorts of histology slides of patients at BWH; J. Wang, M. Barbieri, K. Bronstein, L. Cirelli and E. Askeland for querying the BWH slide database and retrieving archival slides; C. Li for assistance with EMRs and Research Patient Data Registry (RPDR); M. Bragg, T. Mellen, T. A. Mages and S. Zimmet for administrative support; Z. Noor for developing the interactive demo website; and K. Tung of Boston Children’s Hospital for anatomical illustrations. This work was supported in part by internal funds from BWH Pathology, NIH NIGMS R35GM138216 (F.M.), Google Cloud Research Grant and Nvidia GPU Grant Program. M.S. was additionally supported by the NIH Biomedical Informatics and Data Science Research Training Program, NIH NLM T15LM007092. The content is solely the responsibility of the authors and does not reflect the official views of the National Institute of Health, National Institute of General Medical Sciences or the National Library of Medicine.
Author information
Authors and Affiliations
Contributions
F.M. and M.Y.L. conceived the study and designed the experiments. M.Y.L. performed the experimental analysis. D.F.K.W., T.Y.C. and M.Z. curated the in-house, external and CUP datasets. M.Y.L., F.M., D.F.K.W., T.Y.C. and M.S. analysed the results. M.Y.L., M.S., J.L. and F.M. developed the data-visualization tools. M.Y.L. and F.M. prepared the manuscript with input from all co-authors. F.M. supervised the research.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Beatrice Knudsen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Overall study design.
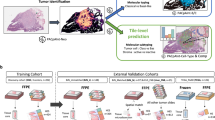
The model was first trained and tested on tumours of known primary origins. For model development and testing, we collected, in total, 32,537 H&E digitized diagnostic slides (from 29,107 patients) with confirmed diagnosis and randomly sampled 70% of cases (22,833 slides) to train the model and 20% of cases (6,499 slides) were held-out for evaluation. The remaining 10% of cases (3,205 slides) was used for validation during training to select the best performing model. To further assess the ability of the model to generalize on data from sources and staining protocols that it did not encounter during training, we also evaluated the model on an external test cohort of 682 cases, submitted from more than 200 US and international medical centres. The model was then assesed on increasingly difficult cases of metastatic tumours. Lastly, to assess the ability of the model to inform meaningful predictions for origins of cancers that cannot be readily diagnosed by human experts using H&E histology alone, we curated an additional diverse dataset of 743 cases of CUP sourced from institutions across the country and outside the USA. Although the primary cancer could not be initially assigned for all of these cases based on H&E histology alone, using EMRs and evidence from clinical and ancillary tests, we identified a subset of 317 cases for which a primary differential was eventually assigned over the course of the patient’s history (see Methods). We validated our model against the recorded primary differential for agreement, showcasing the applicability of the model to cases without clear morphological indication for a particular primary cancer.
Extended Data Fig. 2 Classification performance for the prediction of cancer origins on metastatic tumours.
a, The confusion matrix, along with the precision and recall of each class and its count is plotted for metastatic tumours in the test set (n = 1,408). Glioma was excluded as there were no metastatic glioma specimens in the test set and it was verified that no case of metastasis was predicted as glioma by the model. b. The micro-averaged, one-versus-rest AUC ROC. c, Top-k accuracies of the model on only metastatic tumours (n = 1,408), and on the combined set of metastatic and primary tumours (n = 6,499). d, Accuracy of the model on metastatic tumours binned into different levels of prediction confidence. a, c, d, Error bars indicate 95% confidence intervals, the centre is always the computed value of each classification performance metric (specified by its respective axis labels).
Extended Data Fig. 3 Performance for the prediction of cancer origins on metastatic and primary tumours.
a, b, Additional metrics including per-class and micro-averaged F1-score and mean average precision score are computed for the combined set of primary and metastatic tumours (a; n = 6,499) and only metastatic tumours (b; n = 1,408) in the test set. a, b, Error bars indicate 95% confidence intervals, plotted around the computed value of each classification performance metric (specified by its respective axis labels). Note that the micro-averaged F1-score is the same as the overall accuracy. See Supplementary Table 3 for the number of metastatic and primary tumours for each origin in the test set.
Extended Data Fig. 4 Ablation studies.
a, b, Ablation experiments were performed to assess the benefit of multitask learning and including patient sex as an input in addition to histology on the performance for the prediction of cancer origins (see Methods, ‘Ablation studies’). Top-k accuracies for testing on both primary and metastatic tumours (a; n = 6,499) in the held-out test set and testing on only metastatic tumours (b; n = 1,408). The multitask model with access to patient sex scored nearly 2.0% higher in top-1 accuracy compared to the baseline, single-task model using histology only when testing on the entire test set, and is 6.8% higher when testing on only the metastatic tumours. c, Additional experiments are performed to assess the importance of including primary tumour slides during training and the effect of adding the tissue sampling or biopsy site as another input covariate (in addition to sex) on model performance on metastatic tumours (n = 1,408). The accuracy of the model decreased by 8.5% when trained on only metastatic tumours in the training set, showing that the ability of the model to recognize metastatic tumours benefits substantially from also learning from primary tumours. We additionally experimented with providing the tissue sampling or biopsy site to the model. Multitask training is used when training on both primary and metastatic tumours. A decrease of 4.6% in model accuracy is observed when the biopsy site information is incorporated. This is probably because the biopsy site can provide a direct shortcut to the ground truth label for primary tumour slides and therefore discourages the model from learning from the morphology of primary tumours, which we have found to be beneficial for the ability of the model to recognize metastatic tumours. a–c, Error bars indicate 95% confidence intervals, plotted around the computed value of each classification performance metric (specified by its respective axis labels).
Extended Data Fig. 5 Model performance on the binary problem of distinguishing between primary and metastatic tumours.
a, Performance for tumours at common metastatic sites. The AUC ROCs (y axis) with associated 95% confidence intervals and ROC curves are shown for organ sites (x axis) with at least 10 metastatic and 10 primary tumours in the test set. The ovary, uterus and cervix were grouped into upper female reproductive tract (‘Müllerian’). The number of primary tumours (first element) and metastatic tumours (second element) at each site are indicated as a tuple above each bar. b, Performance for tumours of different primary origins. The AUC ROCs (y axis) with associated 95% confidence intervals and ROC curves are shown for tumours from each origin site (x axis) except for glioma, for which no metastatic tumours were present in our test set. The number of primary tumours (first element) and metastatic tumours (second element) for each origin are indicated as a tuple above each bar. a, b, Without the loss of generality, metastatic tumours are designated as the ‘positive’ class, and primary tumours as the ‘negative’ class for computing sensitivities and specificities. The operating point of the model is indicated by a red dot on each ROC curve, and is based on maximizing Youden’s J index.
Extended Data Fig. 6 Model performance on difficult metastatic and unknown primary tumours.
a, The performance of the model for the prediction of cancer origins is evaluated in terms of top-k accuracies (acc) and Cohen’s κ score for patients with metastatic tumours in the held-out test set (n = 1,408). Performance is additionally reported for subsets of patients with metastatic tumours depending on the number of diagnostic IHC stains used, whether recommendation for clinical or radiological correlation was given and whether the tumour was categorized as poorly differentiated. b, For the held-out test set of cases of CUP with assigned primary differential diagnosis (n = 317), the model performance is assessed using agreement (agr) with the assigned differential. Performance is additionally reported for high-confidence model predictions (for example, model confidence ≥ 0.5) as well as for cases with a high versus low degree of diagnostic certainty associated with the assigned differential. For cases of CUP, based on the strength of evidence used to support the differential diagnosis and language used in EMRs, we define high-certainty diagnoses as being compatible with morphological evidence or supported by IHC findings or clinical, radiological or molecular correlation, whereas low-certainty diagnoses may not suggest a single specific primary origin or lacked definitive supporting evidence for the assigned primary differential. a, b, Error bars indicate 95% confidence intervals, plotted around the computed value of each classification performance metric (specified by its respective axis labels).
Extended Data Fig. 7 Examples of metastases from colorectal, breast and lung primary tumours with attention heat maps.
a–c, Example metastases from colorectal (a), breast (b) and lung (c) primary tumours are shown. For each case, the attention heat map of the model is displayed on top of the original H&E WSI as a semi-transparent overlay in which the overlaid regions range from crimson (high attention, high diagnostic relevance) to navy (low attention, low diagnostic relevance). Left, sites of metastasis are shown, including the lung, lymph node (LN), liver and brain. Right, H&E images show, from left to right, low magnification with corresponding attention map, medium magnification with corresponding attention map, and high-magnification patches. a, Medium- and high-magnification views demonstrate so-called ‘dirty necrosis’ and variably sized glands with densely packed, hyperchromatic nuclei that are characteristic of colorectal adenocarcinoma. b, Medium- and high-magnification views demonstrate sheets of cells as well as small tubules and glands—morphologies that are consistent with metastatic breast carcinomas. c, Medium- and high-magnification views demonstrate sheets of cells, variably sized glands and cells in infiltrative single files. The cells have large, hyperchromatic nuclei and high nuclear:cytoplasmic ratios, which are consistent with metastatic lung carcinomas. a–c, The attention heat maps allow the predictions of the model for each case to be visually interpretable for human experts, revealing the morphological features used by the model for the determination of the classification. High-resolution heat maps for cases from all primary sites can be accessed through our interactive demo website (http://toad.mahmoodlab.org).
Extended Data Fig. 8 TOAD-assisted CUP work-up: example 1.
Top, a representative case that underwent a standard CUP work-up involving extensive IHC staining and clinical correlation. Strong PAX8 staining suggested a Müllerian origin and multiple IHC tests were used to rule out other primary tumours. Retrospectively, we analysed the case with TOAD and found that the top-3 determinations were ovarian, breast and lung, and, after this determination, that only three IHC stains (PAX8, GATA3 and TTF1) needed to be used to confirm a Müllerian origin and rule out breast carcinoma and lung adenocarcinoma. This workflow demonstrates how TOAD can be used as an assistive diagnostic tool. Bottom, medium magnification and corresponding heat maps of representative areas of tumour, with high-magnification, high-attention patches on the right outlined in crimson and low-attention patches outlined in navy.
Extended Data Fig. 9 Analysis of high-attention regions for metastatic tumours.
Relative counts of different cell types localized within the high-attention regions proposed by the model were quantified. Specifically, the top-10 high-attention patches from each slide were extracted at the 20× equivalent magnification and a HoverNet35 model trained for multi-organ nucleus segmentation and classification was used to detect different cellular populations including tumour cells (red), lymphocytes (green), connective tissue (blue), dead cells (yellow) and non-neoplastic epithelial cells (orange). The fraction of cells for each cell type is plotted using box plots for all metastatic slides in the test set (n = 1,408) and is stratified by each primary origin site: lung (n = 236), breast (n = 231), colorectal (n = 175), pancreatobiliary (n = 122), skin (n = 111), ovarian (n = 102), renal (n = 79), prostate (n = 64), head and neck (n = 57), oesophagogastric (n = 52), thyroid (n = 43), bladder (n = 42), germ cell (n = 32), endometrial (n = 21), liver (n = 18), adrenal (n = 12) and cervix (n = 11). Boxes indicate quartile values and whiskers extend to data points within 1.5× the interquartile range. This analysis demonstrates in addition to the attention heat maps, that the model attends strongly to regions of tumour presence for its predictions.
Extended Data Fig. 10 Classification performance of adenocarcinoma network, squamous cell carcinoma network and site-specific networks for tumour metastasized to the liver and lymph node.
a, b, Often pathologists can readily distinguish between adenocarcinoma and squamous cell carcinoma based on the morphological and architectural appearance of the tumour cells that are present in the tissue. However, within the respective family of adenocarcinoma and squamous cell carcinoma subtypes, determining the origin of the tumour can remain a challenging task. Therefore, we hypothesized that we can develop models to specifically predict the origin of tumours for top primary sites of adenocarcinoma (a) and squamous cell carcinoma (b). Cases from six primary sites (breast, lung, colorectal, pancreatobiliary, prostate and oesophagogastric) and four primary sites (head and neck, lung, cervix and oesophagogastric) were chosen for the development of the adenocarcinoma and squamous cell carcinoma classifiers, respectively, based on their frequency in the database. We also explored the additional scenarios of predicting the primary origins of metastatic tumours grouped by a common metastatic site, including the liver (c) and lymph node (d). Cases of metastasis from the top-four and top-seven primary origins for liver and lymph nodes, respectively, were chosen on the basis of their frequency in our database. See Methods, ‘Additional experiments and analysis’ for details. a–d, Left, the confusion matrix, along with the precision and recall of each class and its count is plotted for the adenocarcinoma model test set (a; n = 2,920) and squamous cell carcinoma model test set (b; n = 621), the liver metastasis (met.) site test set (c; n = 223) and lymph node metastasis site test set (d; n = 318), respectively. Consistent with the model developed using examples of all 18 primary sites, the adenocarcinoma-, squamous-cell-carcinoma- and site-specific models were trained by including the sex of the patient. Performance for models trained with and without the sex of the patient in terms of the micro-averaged, one-versus-rest AUC ROC (middle) and F1-scores for each primary site and overall model accuracy (micro-averaged F1-score) (right) are shown. All error bars indicate 95% confidence intervals, plotted around the computed value of each classification performance metric (specified by its respective axis labels).
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-6, Supplementary Tables 1, 3, 6, 7, 11, 13 and 14, and legends for Supplementary Tables 2, 4, 5, 8, 9, 10 and 12 (see separate Excel file for these Tables).
Supplementary Tables
This file contains Supplementary Tables 2, 4, 5, 8, 9, 10 and 12.
Rights and permissions
About this article
Cite this article
Lu, M.Y., Chen, T.Y., Williamson, D.F.K. et al. AI-based pathology predicts origins for cancers of unknown primary. Nature 594, 106–110 (2021). https://doi.org/10.1038/s41586-021-03512-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03512-4
This article is cited by
-
Applying the UTAUT2 framework to patients’ attitudes toward healthcare task shifting with artificial intelligence
BMC Health Services Research (2024)
-
Performance and application of the total-body PET/CT scanner: a literature review
EJNMMI Research (2024)
-
DeepRisk network: an AI-based tool for digital pathology signature and treatment responsiveness of gastric cancer using whole-slide images
Journal of Translational Medicine (2024)
-
Predicting 5-year recurrence risk in colorectal cancer: development and validation of a histology-based deep learning approach
British Journal of Cancer (2024)
-
A population-level digital histologic biomarker for enhanced prognosis of invasive breast cancer
Nature Medicine (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.