Abstract
The accurate segregation of chromosomes during meiosis—which is critical for genome stability across sexual cycles—relies on homologous recombination initiated by DNA double-strand breaks (DSBs) made by the Spo11 protein1,2. The formation of DSBs is regulated and tied to the elaboration of large-scale chromosome structures3,4,5, but the protein assemblies that execute and control DNA breakage are poorly understood. Here we address this through the molecular characterization of Saccharomyces cerevisiae RMM (Rec114, Mei4 and Mer2) proteins—essential, conserved components of the DSB machinery2. Each subcomplex of Rec114–Mei4 (a 2:1 heterotrimer) or Mer2 (a coiled-coil-containing homotetramer) is monodispersed in solution, but they independently condense with DNA into reversible nucleoprotein clusters that share properties with phase-separated systems. Multivalent interactions drive this condensation. Mutations that weaken protein–DNA interactions strongly disrupt both condensate formation and DSBs in vivo, and thus these processes are highly correlated. In vitro, condensates fuse into mixed RMM clusters that further recruit Spo11 complexes. Our data show how the DSB machinery self-assembles on chromosome axes to create centres of DSB activity. We propose that multilayered control of Spo11 arises from the recruitment of regulatory components and modulation of the biophysical properties of the condensates.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Processed crosslinking-mass spectrometry data are provided in Supplementary Table 1. Fasta sequences of the yeast SK1 strain are available at https://www.yeastgenome.org. Swissprot reviewed database is available at https://www.uniprot.org/. Source data are provided with this paper.
Code availability
The custom ImageJ scripts for analysis of condensate foci are available at https://github.com/claeysbouuaert/scripts.
References
de Massy, B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47, 563–599 (2013).
Lam, I. & Keeney, S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb. Perspect. Biol. 7, a016634 (2014).
Cooper, T. J., Garcia, V. & Neale, M. J. Meiotic DSB patterning: a multifaceted process. Cell Cycle 15, 13–21 (2016).
Keeney, S., Lange, J. & Mohibullah, N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu. Rev. Genet. 48, 187–214 (2014).
Kleckner, N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115, 175–194 (2006).
Keeney, S., Giroux, C. N. & Kleckner, N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997).
Bergerat, A. et al. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature 386, 414–417 (1997).
Robert, T. et al. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science 351, 943–949 (2016).
Vrielynck, N. et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science 351, 939–943 (2016).
Claeys Bouuaert, C. et al. Structural and functional characterization of the Spo11 core complex. Nat. Struct. Mol. Biol. 28, 92–102 (2021).
Li, J., Hooker, G. W. & Roeder, G. S. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 173, 1969–1981 (2006).
Maleki, S., Neale, M. J., Arora, C., Henderson, K. A. & Keeney, S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma 116, 471–486 (2007).
Steiner, S., Kohli, J. & Ludin, K. Functional interactions among members of the meiotic initiation complex in fission yeast. Curr. Genet. 56, 237–249 (2010).
Miyoshi, T. et al. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol. Cell 47, 722–733 (2012).
Henderson, K. A., Kee, K., Maleki, S., Santini, P. A. & Keeney, S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell 125, 1321–1332 (2006).
Panizza, S. et al. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146, 372–383 (2011).
Arora, C., Kee, K., Maleki, S. & Keeney, S. Antiviral protein Ski8 is a direct partner of Spo11 in meiotic DNA break formation, independent of its cytoplasmic role in RNA metabolism. Mol. Cell 13, 549–559 (2004).
Sommermeyer, V., Béneut, C., Chaplais, E., Serrentino, M. E. & Borde, V. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol. Cell 49, 43–54 (2013).
Acquaviva, L. et al. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 339, 215–218 (2013).
Kumar, R. et al. MEI4 — a central player in the regulation of meiotic DNA double-strand break formation in the mouse. J. Cell Sci. 128, 1800–1811 (2015).
Stanzione, M. et al. Meiotic DNA break formation requires the unsynapsed chromosome axis-binding protein IHO1 (CCDC36) in mice. Nat. Cell Biol. 18, 1208–1220 (2016).
Robert, T., Vrielynck, N., Mézard, C., de Massy, B. & Grelon, M. A new light on the meiotic DSB catalytic complex. Semin. Cell Dev. Biol. 54, 165–176 (2016).
Tessé, S. et al. Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev. 31, 1880–1893 (2017).
Wang, W. et al. Homozygous mutations in REC114 cause female infertility characterised by multiple pronuclei formation and early embryonic arrest. J. Med. Genet. 57, 187–194 (2020).
Kumar, R. et al. Mouse REC114 is essential for meiotic DNA double-strand break formation and forms a complex with MEI4. Life Sci. Alliance 1, e201800259 (2018).
Kumar, R., Bourbon, H. M. & de Massy, B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 24, 1266–1280 (2010).
Boekhout, M. et al. REC114 partner ANKRD31 controls number, timing, and location of meiotic DNA breaks. Mol. Cell 74, 1053–1068.e8 (2019).
Engebrecht, J. A., Voelkel-Meiman, K. & Roeder, G. S. Meiosis-specific RNA splicing in yeast. Cell 66, 1257–1268 (1991).
Lorenz, A., Estreicher, A., Kohli, J. & Loidl, J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma 115, 330–340 (2006).
Bonfils, S., Rozalén, A. E., Smith, G. R., Moreno, S. & Martín-Castellanos, C. Functional interactions of Rec24, the fission yeast ortholog of mouse Mei4, with the meiotic recombination-initiation complex. J. Cell Sci. 124, 1328–1338 (2011).
Li, P. et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340 (2012).
Wheeler, J. R., Matheny, T., Jain, S., Abrisch, R. & Parker, R. Distinct stages in stress granule assembly and disassembly. eLife 5, e18413 (2016).
Su, X. et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352, 595–599 (2016).
Boulay, G. et al. Cancer-specific retargeting of BAF complexes by a prion-like domain. Cell 171, 163–178.e19 (2017).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Boeynaems, S. et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol. 28, 420–435 (2018).
Lin, Y., Protter, D. S., Rosen, M. K. & Parker, R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell 60, 208–219 (2015).
Patel, A. et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015).
Xiang, S. et al. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 163, 829–839 (2015).
Garcia, V., Gray, S., Allison, R. M., Cooper, T. J. & Neale, M. J. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 520, 114–118 (2015).
Johnson, D. et al. Concerted cutting by Spo11 illuminates the mechanism of meiotic DNA break formation. Nature (in the press).
Dosztányi, Z. Prediction of protein disorder based on IUPred. Protein Sci. 27, 331–340 (2018).
Sebastiaan Winkler, G. et al. Isolation and mass spectrometry of transcription factor complexes. Methods 26, 260–269 (2002).
Erdjument-Bromage, H. et al. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J. Chromatogr. A 826, 167–181 (1998).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Combe, C. W., Fischer, L. & Rappsilber, J. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteomics 14, 1137–1147 (2015).
Nesvizhskii, A. I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Murakami, H., Borde, V., Nicolas, A. & Keeney, S. Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions. Methods Mol. Biol. 557, 117–142 (2009).
Neale, M. J. & Keeney, S. End-labeling and analysis of Spo11-oligonucleotide complexes in Saccharomyces cerevisiae. Methods Mol. Biol. 557, 183–195 (2009).
Acknowledgements
We thank A. Nicolas and V. Borde for sharing unpublished information; J. Xu for assistance with preliminary analyses of the Mer2(KRRR) mutant; other members of the Keeney lab for discussions; MSK core facilities, supported by NIH cancer center core grant P30 CA008748: Microchemistry and Proteomics (R. Hendrickson and E. Chang) for the XL–MS experiments; Molecular Cytology (M. Brendel and Y. Romin) for AFM experiments; S. Fujisawa for writing a Fiji script to quantify fluorescent foci; and E. Folta-Stogniew from the Biophysics Resource of Keck Facility at Yale University for the SEC-MALS experiments. The SEC–LS/UV/RI instrumentation was supported by NIH grant S10 RR023748. This work was supported by the Howard Hughes Medical Institute (S.K.), the Maloris Foundation (D.P.), an MSK Basic Research Innovation Award (S.K. and D.P.), the European Research Council under the European Union’s Horizon 2020 research and innovation program (ERC grant agreement 802525 to C.C.B.), and the Fonds National de la Recherche Scientifique (MIS-Ulysse grant F.6002.20 to C.C.B.).
Author information
Authors and Affiliations
Contributions
C.C.B. and S.K. designed the study and supervised the research; C.C.B. carried out all experiments except as noted; S.P. performed yeast two-hybrid experiments (Fig. 4g, Extended Data Figs. 1k, 8e) and assisted C.C.B. with the generation of expression constructs, virus preparation and protein purification; J.W. performed SEC–MALS analyses of mutant protein constructs (Fig. 1j, Extended Data Fig. 1h, j) and W.X. performed FRAP experiments (Extended Data Fig. 5c) under the supervision of D.J.P.; C.O. performed MBP pulldown (Extended Data Fig. 7d) and trypsin proteolysis experiments (Extended Data Fig. 6h) and D.D. performed the condensate mixing experiments (Extended Data Figs. 6e, 8f) under the supervision of C.C.B.; C.C.B. and S.K. wrote the paper with input from the other authors; C.C.B., D.J.P. and S.K. secured funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of the Rec114–Mei4 complex.
a, Strategy for purification of a hypothetical Rec114–Mei4–Mer2 (RMM) complex. Combinations of MBP-tagged and HisFlag-tagged RMM subunits were co-expressed in insect cells. After cell lysis, complexes were purified by sequential affinity chromatography and analysed by SDS–PAGE. Expression and solubility of the recombinant proteins were verified by western blotting (WB) of cell extracts. b, Analysis of purified complexes. Rec114–Mei4 complexes were apparent (lanes 1 and 3), but no Mer2 was co-purified. Lanes 2 and 4 show some enrichment of MBP–Mer2, but no co-purification of Rec114–Mei4. The presence of MBP–Mer2 in lanes 2 and 4 of the silver-stained gel may be due to background binding of MBP–Mer2 to the NiNTA resin (potentially via adsorption of DNA to the resin), or to low-affinity interactions with immobilized His-tagged Rec114–Mei4 complexes. Either way, none of the combinations tested yielded stoichiometric complexes of all three RMM subunits. Western blot controls of cell extracts showed that the tagged RMM proteins were expressed and soluble. c, Mass spectrometry analysis of Rec114–Mei4 complexes. Purified Rec114–Mei4 complexes were treated with trypsin and analysed by LC–MS/MS. The ratio of spectral counts between Rec114 and Mei4 provides additional evidence supporting the 2:1 stoichiometry of the complex. d, e, Alignments and predicted secondary structures of the C terminus of Rec114 (d) and the N terminus of Mei4 (e). The positions of the conserved SSMs are indicated. f, Cartoon of the Rec114–Mei4 truncations analysed. g, Purification of Rec114–Mei4 truncations. Proteins were expressed in E. coli and purified on NiNTA resin using a HisSUMO tag fused to the N terminus of the Rec114 fragment. After removal of the tag by treatment with the SUMO protease Ulp1, complexes were further purified by gel filtration. A Coomassie-stained SDS–PAGE analysis of purified complexes is shown (5 μg was loaded for each sample). Polypeptides containing Rec114(375–428) and Mei4(1–43) retained the ability to interact (combination #4). h, SEC–MALS analysis of Rec114–Mei4 truncations. The data are consistent with expectations for truncations that contain two Rec114 subunits and one Mei4 subunit. The C terminus of Rec114 alone forms a dimer. i, Wild-type and F411A-containing variants of HisSUMO–Rec114(325–428) were co-expressed with Mei4(1–90) and purified by chromatography on NiNTA resin. The absence of the Mei4 fragment with Rec114(F411A) shows that the mutation abolishes the interaction with Mei4. j, SEC–MALS analysis of untagged wild-type (WT, reproduced from h to aid comparison) and F411A mutant Rec114(325–428) shows that the mutation affects Rec114 dimerization. k, Y2H analysis of the interaction of Gal4BD–Rec114 (wild-type and F411A) with LexA–Mei4, LexA–Rec102, or LexA–Rec104 (mean + s.d. from four replicates). β-Gal units are quantified by hydrolysis of ONPG. The F411A mutation abolishes the interaction of Rec114 with Mei4, but not with Rec102 or Rec104. l, Southern blot analysis of meiotic DSB formation at the CCT6 hotspot, showing that the rec114F411A strain is defective in meiotic DSB formation. m, Spore viability of rec114F411A mutant (n = 40). n, Western blot analyses of meiotic protein extracts from myc-tagged wild-type and rec114F411A strains. The F411A mutation does not compromise the expression of Rec114. o, Left, immunofluorescence microscopy analysis of meiotic chromosome spreads with wild-type and F411A myc-tagged Rec114. Green, anti-myc; red, synaptonemal complex component Zip1; blue, DNA. Right, number of Rec114 foci per leptotene or early zygotene cell; mean ± s.d. (n = 20 and 38 cells for wild-type and F411A, respectively). The F411A mutation abolishes the formation of chromatin-associated Rec114 foci.
Extended Data Fig. 2 DNA-binding properties of Rec114–Mei4 and Mer2 complexes.
a, b, Gel shift analysis of binding of Rec114–Mei4 (a) and Mer2 (b) to 20- or 40-bp DNA substrates. Quantification is in Fig. 2b. c, d, Competition assay of Rec114–Mei4 (c) or Mer2 (d) binding to 80 bp radiolabelled DNA (1 nM) in the presence of 20 or 80 bp cold competitor. Fold excess is in nucleotides. Lines are one-phase decay fits. e, f, Binding to plasmid DNA analysed by native agarose gel electrophoresis. Rec114–Mei4 (e) and Mer2 (f) were titrated with 2 nM plasmid DNA (pUC19) in the presence or absence of 5 mM MgCl2. Rec114–Mei4 complexes bound with roughly similar affinity independently of the presence of Mg2+ (apparent KD ≈ 50–80 nM). The apparent affinity is substantially lower than suggested by the gel shift analyses with radiolabelled substrates presented in a and Fig. 2a, b (apparent affinities in Fig. 2 legend). We suggest that this difference is because the proteins coalesce on a small fraction of the plasmid molecules, as illustrated in the cartoon below. Indeed, bound plasmids remained trapped in the wells, which is consistent with cooperative assembly of large nucleoprotein structures. Because each plasmid substrate provides many more binding sites than the short oligo substrates in a and Fig. 2a, a higher concentration of protein is required to reach complete binding of all of the plasmid molecules. In contrast to Rec114–Mei4, Mer2 showed efficient binding in the absence of Mg2+ in this assay (KD = 30 ± 2 nM) but binding appeared to be considerably inhibited in the presence of Mg2+ (KD ≈ 150 nM), as indicated by the persistence of unbound substrate at high protein concentrations. However, although the electrophoretic mobility of Mer2-bound plasmids decreased steadily as the concentration of Mer2 increased in the absence of Mg2+, no such steady progression was observed when Mg2+ was included. Instead, a minority of bound substrates shifted to a low-mobility species (* in f), indicating that they were occupied by multiple Mer2 complexes. We interpret this as that, rather than inhibiting DNA binding, Mg2+ promotes cooperativity, in agreement with the fluorescence microscopy analysis (Extended Data Fig. 3b). The difference in migration distance of the plasmid between the gels with and without Mg2+ is due to the presence of Mg2+ in the electrophoresis buffer. g, AFM imaging of 12 nM Rec114–Mei4 in the absence (left) or presence (right) of 1 nM plasmid DNA (pUC19).
Extended Data Fig. 3 Properties of Rec114–Mei4 and Mer2 DNA-dependent condensates.
a, b, Visualization of nucleoprotein condensates by epifluorescence microscopy using tagged Rec114–Mei4 (a) or Mer2 (b) in the presence or absence of 5 mM MgCl2. Foci were defined using a fixed intensity threshold between samples. Each point represents the measurement from a field of view. Mean ± s.d. from 10 fields of view of 1.7 × 104 μm2 (a) or 27 and 26 sections of 400 μm2 with and without Mg2+, respectively (b). c–f, Effect of fluorophore labelling or tagging on the DNA-binding and DNA-driven condensation activities of Rec114–Mei4 and Mer2 complexes. Labelling with Alexa594 or Alexa488 was achieved using amine-reactive fluorophores. Tagging was achieved by fusion of Rec114 with the monomeric fluorescent protein mScarlet or fusion of Mer2 with the weakly dimerizing fluorescent protein eGFP. The results described here indicate that covalent Alexa labelling has little, if any, effect on the DNA binding properties of these complexes, whereas fluorescent protein tagging caused subtle alterations in DNA binding and/or condensation. In most subsequent experiments, we used the dye-labelled complexes to minimize steric effects or oligomerization of fluorescent protein tags. c, Gel-shift analysis of binding of unlabelled, Alexa594-labelled, or mScarlet-tagged Rec114–Mei4 complexes to an 80-bp radiolabelled DNA substrate. The three versions of the Rec114–Mei4 complex have the same intrinsic DNA-binding activity. d, Gel-shift analysis of binding of unlabelled, Alexa488-labelled, or eGFP-tagged Mer2 complexes to an 80-bp radiolabelled DNA substrate. The DNA-binding activity of the Alexa-labelled Mer2 complex is nearly identical to the untagged protein, but the eGFP-tagged complex has 3.5-fold reduced DNA-binding activity. e, A comparison between Alexa-labelled and mScarlet-tagged Rec114–Mei4 complexes for DNA-driven condensation. Focus numbers (left) and total fluorescence intensity within foci normalized to the no-PEG samples (right) are shown for the complexes in the presence or absence of 5% PEG. With and without PEG, mScarlet-tagged Rec114–Mei4 produced more foci than the Alexa-labelled version. Because intrinsic DNA binding was indistinguishable between the complexes (c), we infer that the mScarlet-tagged complexes had a reduced efficiency in the cooperative formation of large condensates compared to the Alexa-labelled version, producing more numerous foci. *P < 0.0001 (two-tailed t-test). Mean ± s.d. from 8–10 fields of view. f, As in e, for comparison between Alexa-labelled and eGFP-tagged Mer2 complexes for DNA-driven condensation. The two labelled complexes show different numbers and intensities of foci in the presence of PEG. It is likely that the DNA-binding defect of the eGFP construct (d) leads to the formation of fewer, brighter condensates. It is possible that the weak dimerization activity of eGFP also contributes. *P < 0.0001 (two-tailed t-test). Mean ± s.d. from 9–10 fields of view. g, h, Top, effect of a crowding agent (PEG) on formation of nucleoprotein condensates visualized using covalently fluorophore-labelled Rec114–Mei4 (g) or Mer2 (h). Bottom, effect of protein concentration on DNA-driven condensation in the presence or absence of 5% PEG. Left, focus numbers; right, total fluorescence intensity within foci (normalized to the mean of the highest intensity sample). Mean ± s.d. from 4–6 fields of view (g) or 7–10 fields of view (h). The titrations reveal complex behaviours. g, In the presence of PEG, titration of Rec114–Mei4 from 4 to 32 nM led to a steady decrease in the number of foci, which was accompanied by a concomitant increase in focus intensity. In the absence of PEG, however, the number of Rec114–Mei4 foci first peaked at 8 nM before decreasing as the intensity of the foci started to increase. Nevertheless, focus intensity plateaued at a much lower intensity than in the presence of PEG. h, In the case of Mer2, titration from 25 to 300 nM in the presence of PEG yielded a peak in the number of foci at about 100 nM, which then sharply declined and stabilized beyond 150 nM. Consistently, Mer2 foci remained at a constant, low intensity between 25 and 100 nM, then became abruptly brighter above 100 nM. In the absence of PEG, the number of Mer2 foci increased between 25 and 200 nM, then started to decrease beyond that threshold. These behaviours are likely to reflect the complex combined effects of nucleation, growth, and collapse of the condensates, each of which is each affected differently by protein concentrations and by the crowding effect provided by PEG. See Source Data for exact n values for e–h.
Extended Data Fig. 4 Properties of Rec114–Mei4 and Mer2 DNA-dependent condensates.
a, b, Effect of challenging Rec114–Mei4 (a) or Mer2 (b) nucleoprotein condensates with DNase I or 0.5 M NaCl. Condensates were assembled for 5 min before challenge. Bottom, quantification of focus numbers per 1,000 μm2 and of the total fluorescence intensity within foci within fields of view (normalized to mean of the no-treatment controls). Mean ± s.d. from 5–10 fields of view. c, e, Titrations of Rec114–Mei4 (c) and Mer2 (e) in the presence of DNA and PEG and various concentrations of NaCl. Heat maps represent the fraction of fluorescence signal found within foci. Condensed fractions are maximal at high protein and low salt concentrations. At all protein concentrations, condensation is essentially abolished beyond 250 mM NaCl. This suggests that electrostatic interactions, probably between the negatively charged DNA backbone and positively charged protein residues, are important for condensation. d, f, Time dependence for irreversibility of Rec114–Mei4 (d) and Mer2 (f) condensates. Some phase-separated liquid droplets have been shown to mature over time and progressively adopt gel-like or solid states35,37,38,39. Such sol–gel transitions may occur spontaneously through different mechanisms, including fibrillization and entanglement, and are thought to be counteracted in vivo to prevent the progressive accumulation of amyloid-like structures associated with pathological states35. To address whether our condensates are prone to progressive hardening, we queried the effect of assembly time on reversibility. We performed a time-course experiment in which the condensates were challenged by treatment with 0.5 M NaCl after an indicated period of assembly in the presence or absence of PEG. The graphs show the total intensity summed for foci within fields of view, expressed as a percentage of the intensity without a salt challenge. Mean ± s.d. for 6–10 fields of view. With Rec114–Mei4, 10% and 50% of fluorescent signal became refractory to the salt wash within 5 min of incubation time in the absence and presence of PEG, respectively (see a for example images and quantification). With Mer2, there was no evidence for the formation of irreversible structures in the absence of PEG during the course of the experiment. However, up to 25% of the focus intensity resisted the salt wash treatment after 8 min of incubation time in the presence of PEG. Therefore, both Rec114–Mei4 and Mer2 have a propensity to form more stable, perhaps gel-like, structures over time. Under our experimental conditions, this was more evident for Rec114–Mei4 than for Mer2, and was accentuated by molecular crowding. g, h, Assembly of Rec114–Mei4 (g) and Mer2 (h) with fluorescently labelled 9.6-kb and 100-bp linear DNA substrates. The overlap between the protein foci and puncta of DNA shows that the DNA is also enriched in the condensates. However, in contrast to the protein signal, the fluorescent signal of the DNA covers the slide because DNA is in excess and does not condense by itself. i, j, Competition between long and short DNA substrates for incorporation into condensates. Rec114–Mei4 (i) or Mer2 (j) condensates were assembled in the presence of a fluorescently labelled DNA substrate with or without 20-fold nucleotide excess of unlabelled competitor. The amount of fluorescent DNA signal averaged between ten foci is plotted. In each case, the 9.6-kb substrate was a more effective competitor than the 100-bp substrate. In addition, the 100-bp substrate was more successful at competing with the 100-bp fluorescent substrate than with the 9.6-kb fluorescent substrate. This preference for large DNA substrates is consistent with the hypothesis that the condensates form through multivalent interactions between the positively charged residues of Rec114–Mei4 or Mer2 and the sugar-phosphate backbone of the DNA. See Source Data for exact n values for a, b, d, f.
Extended Data Fig. 5 Growth of DNA-driven condensates by fusion.
a, Three scenarios for the assembly of DNA-driven condensates. (i) Nucleation could be limiting, with focus growth resulting principally from incorporation of protein from soluble pools. (ii) Frequent nucleation events could occur, initially leading to large numbers of small foci, whereupon some foci dissolve and others grow. (iii) Frequent nucleation could yield numerous small foci that then collide and fuse to yield fewer, larger foci. See Supplementary Discussion 2 for more detail. b, Time course of the assembly of Rec114–Mei4 foci in the presence of plasmid DNA. The x-axis indicates the time in solution before plating, upon which DNA is immobilized to the glass slide while soluble protein is still free to diffuse. Plot shows focus numbers and average focus intensity (normalized to the mean at 30 min). Mean ± s.d. from ten fields of view. c, FRAP experiments with Mer2 and Rec114–Mei4 condensates. Mean ± s.d. for six photobleached condensates.
Extended Data Fig. 6 Identification of DNA-binding residues and effect of DNA binding on condensation in vitro and in vivo and on Spo11-induced break formation.
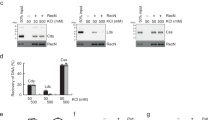
a, Mapping the DNA-binding domain of Rec114–Mei4 complexes. Gel-shift analysis was performed with pUC19 plasmid DNA and the Rec114–Mei4 protein constructs shown in Extended Data Fig. 1f. Constructs #2, #3 and #4, which include the C terminus of Rec114 and the N terminus of Mei4, were competent for DNA binding. The difference in mobility of shifted species between these constructs is in line with the difference in sizes of the protein complexes. Mei4 is dispensable for DNA binding by Rec114 (construct #5 lacks Mei4). The N terminus of Rec114 alone, encompassing the PH domain, did not bind DNA (construct #6). None of the constructs showed evidence for cooperative DNA binding (unlike the full-length protein (Extended Data Fig. 2e)), suggesting that they do not undergo DNA-driven condensation. b, Gel shift analysis of wild-type and mutant Rec114–Mei4 complexes binding to an 80-bp DNA substrate. The Rec114(4KR) mutant has residues R395, K396, K399, and R400 mutated to alanine. Lines on graphs are sigmoidal curve fits. c, Mapping the DNA-binding domain of Mer2. Gel-shift analysis was performed with pUC19 plasmid DNA and HisSUMO-tagged Mer2 protein that was full-length (FL), had the N terminus removed (fragment 77–314), or had both the N and C termini removed (fragment 77–227). Deleting the N terminus alone had no significant effect on DNA binding, but further deleting the C terminus strongly reduced DNA binding. d, Effect of the Rec114(4KR) mutation on condensation in vitro. Reactions included 5% PEG. Each point is the average of the intensities of foci in a field of view (n = 20 fields), normalized to the overall mean for wild type. Mean ± s.d. e, Incorporation of Mer2(KRRR) into preformed condensates. Condensates were assembled with 100 nM unlabelled Mer2. Reactions were then supplemented with the indicated amount eGFP–Mer2 (wild-type or KRRR) and plated immediately. Incorporation of eGFP-tagged complexes within condensates was quantified. Mean ± s.d. from 20 fields of view. f, Immunofluorescence on meiotic chromosome spreads for myc-tagged Rec114. The number of foci per leptotene or early zygotene cell is plotted. Mean ± s.d. (n = 44 and 40 cells for wild-type and rec1144KR strains, respectively). g, Immunoblotting of meiotic protein extracts for wild-type and mutant Rec114 (left) or Mer2 (right). h, Partial proteolysis of wild-type and mutant Mer2 and Rec114–Mei4 complexes. i, Immunoblot analysis of wild-type Mer2 and Mer2(KRRR). Protein extracts of meiotic time courses were analysed by SDS–PAGE followed by immunoblotting against Mer2–myc. Anti-Kar2 was used as a loading control. Quantification of immunoblot signal is plotted. Mer2(KRRR)–myc reached higher steady-state protein levels and persisted longer than wild-type Mer2–myc. A previous study showed that mutating an essential CDK phosphorylation site of Mer2 (Ser30) or inhibiting CDK activity led to reduced turnover of Mer2, similar to the effect of the KRRR mutant15. This is consistent with the hypothesis that Mer2 turnover is tied to phosphorylation, which requires DNA binding. j, Southern blot analysis of meiotic DSB formation at the CCT6 hotspot in strains expressing wild-type or mutant Rec114 protein. k, Labelling of Spo11–oligo complexes in wild-type and mutant Rec114 (left) and Mer2 (right) strains. Points represent two biological replicates. l, Spore viability of strains expressing wild-type or mutant Rec114 (left) and Mer2 (right) (n = 40). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 7 Rec114–Mei4 and Mer2 form mixed condensates.
a, Rec114–Mei4 colocalizes with Mer2 in mixed condensates irrespective of DNA concentration. Reactions containing 16 nM Rec114–Mei4 and 100 nM Mer2 in the presence of 1, 10, or 100 ng μl−1 plasmid DNA were assembled for 20 min at 30 °C. DAPI (5 μg ml−1) was added to the reaction before application to glass slides. DNA enrichment within the condensates is visible at lower DNA concentrations (top, middle), but is not as clear at high DNA concentrations (bottom). The ratios of Rec114–Mei4 (heterotrimers) and Mer2 (tetramers) to each 2.6-kb plasmid DNA molecule are indicated on the right. Colocalization of Rec114–Mei4 and Mer2 complexes is evident even with a molar excess of DNA molecules, demonstrating that formation of joint foci is not simply because both protein complexes are independently associating with a limiting number of DNA substrates. b, Correlated intensity of Rec114–Mei4 and Mer2 proteins within the condensates. Each point shows the fluorescence intensity in an individual focus (n = 950, 925 and 1,000 foci from 2–3 fields of view for samples with 1, 10 and 100 ng μl−1 DNA, respectively), normalized to the average focus intensity per field of view. The strong correlation indicates that the composition of the condensates is highly uniform between foci. In the presence of high DNA concentration, the fraction of smaller foci increased and correlated intensities decreased. c, Recruitment of soluble Rec114–Mei4 (left) or Mer2 (right) into preassembled condensates of Mer2 (left) or Rec114–Mei4 (right). Arrowheads, examples of the preassembled condensates. d, Pulldown of purified Mer2 on amylose resin with or without immobilized Rec114–Mei4 complexes. e, XL–MS of Rec114–Mei4–Mer2 condensates (620 crosslinked peptides, 229 distinct crosslinked pairs of lysines).
Extended Data Fig. 8 Recruitment of the Spo11 core complex to Rec114–Mei4–Mer2 condensates.
a, Quantification of core complex signal within Rec114–Mei4 foci in the presence (100 nM) or absence of Mer2. The average intensity within 20 foci is plotted for each reaction. Shaded areas represent 95% confidence intervals (CI). b, Quantification of core complex signal within Mer2 foci in the presence (16 nM) or absence of Rec114–Mei4. Reactions contained 25 nM Mer2. The average intensity within 20 foci is plotted for each reaction. Shaded areas represent 95% CI. c, Effect of including 100 nM MBP–Rec102–Rec104–HisFlag competitor on the recruitment of the core complex to RMM condensates (16 nM Rec114–Mei4, 100 nM Mer2). The fraction of Rec114–Mei4–Mer2 foci that contain detectable core complex signal is plotted (mean + s.d. from ten fields of view). d, Intensity of core complex signal within Rec114–Mei4–Mer2 condensates in the absence or presence of Rec102–Rec104 competitor. The average core complex intensity within 20 foci is plotted for each reaction. Shaded areas represent 95% CI. e, Mapping regions of Rec114 required for interaction with Rec102 or Rec104 by Y2H analysis. β-Galactosidase units are measured for the interaction between truncated variants of Gal4AD–Rec114 and LexA–Rec102 or LexA–Rec104 (mean + s.d. from four replicates). The position of the HLS mutation within the Rec114 PH-fold is indicated. f, Effect of the HLS mutation on the formation of comingled RMM condensates. Mean ± s.d. from ten fields of view. g, Spore viability of wild-type and rec114HLS mutant strains. h, Immunoblot analysis of meiotic protein extracts from myc-tagged wild-type and rec114HLS mutant strains. Samples from two biological replicates are shown. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 A condensate model for assembly of the meiotic DSB machinery and implications for the control of DSB formation and repair.
a, Assembly of the DSB machinery. Left, Rec114–Mei4 and Mer2 complexes bind DNA in a highly cooperative manner to form large mixed nucleoprotein condensates. Right, these condensates provide a platform to recruit the core complex through interactions that involve the N-terminal domain of Rec114 and the Rec102–Rec104 components of the core complex. Multiple Spo11 complexes are recruited and may engage an incoming DNA loop simultaneously. The molecular arrangement of the core complex proteins is based on ref. 10. See Supplementary Discussion 4 for more detail. b, Hotspot competition and DSB interference. Competition arises before DSB formation as a consequence of the partitioning of RMM proteins into condensates. DSB interference is implemented through local inhibition of further DSB formation by DSB-activated Tel1. Inhibition could work on the same cluster that generated the activation DSB as well as on nearby clusters in cis. See Supplementary Discussion 5 for more detail. c, The coherence provided by the condensates may serve functions during repair, including the maintenance of a physical connection between the DNA ends that involves end-capping by condensate-embedded core complexes. See Supplementary Discussion 6 for more detail.
Supplementary information
Supplementary Information
This file contains Supplementary Discussion, Supplementary References, legend for Supplementary Table 1, Supplementary Tables 2 to 4.
Supplementary Figure 1
This file contains gel source data.
Supplementary Table 1
This file contains XL-MS data.
Rights and permissions
About this article
Cite this article
Claeys Bouuaert, C., Pu, S., Wang, J. et al. DNA-driven condensation assembles the meiotic DNA break machinery. Nature 592, 144–149 (2021). https://doi.org/10.1038/s41586-021-03374-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03374-w
This article is cited by
-
Seeding the meiotic DNA break machinery and initiating recombination on chromosome axes
Nature Communications (2024)
-
Divergence and conservation of the meiotic recombination machinery
Nature Reviews Genetics (2024)
-
Bi-allelic missense variants in MEI4 cause preimplantation embryonic arrest and female infertility
Human Genetics (2024)
-
Recent advances in mechanisms ensuring the pairing, synapsis and segregation of XY chromosomes in mice and humans
Cellular and Molecular Life Sciences (2024)
-
The RNA-binding protein FUS/TLS interacts with SPO11 and PRDM9 and localize at meiotic recombination hotspots
Cellular and Molecular Life Sciences (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.