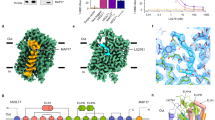
Abstract
Citrate is best known as an intermediate in the tricarboxylic acid cycle of the cell. In addition to this essential role in energy metabolism, the tricarboxylate anion also acts as both a precursor and a regulator of fatty acid synthesis1,2,3. Thus, the rate of fatty acid synthesis correlates directly with the cytosolic concentration of citrate4,5. Liver cells import citrate through the sodium-dependent citrate transporter NaCT (encoded by SLC13A5) and, as a consequence, this protein is a potential target for anti-obesity drugs. Here, to understand the structural basis of its inhibition mechanism, we determined cryo-electron microscopy structures of human NaCT in complexes with citrate or a small-molecule inhibitor. These structures reveal how the inhibitor—which binds to the same site as citrate—arrests the transport cycle of NaCT. The NaCT–inhibitor structure also explains why the compound selectively inhibits NaCT over two homologous human dicarboxylate transporters, and suggests ways to further improve the affinity and selectivity. Finally, the NaCT structures provide a framework for understanding how various mutations abolish the transport activity of NaCT in the brain and thereby cause epilepsy associated with mutations in SLC13A5 in newborns (which is known as SLC13A5-epilepsy)6,7,8.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Electron microscopy densities and protein models have been deposited in the Electron Microscopy Data Bank and Protein Data Bank for the NaCT–citrate (EMD-22457, 7JSK) and NaCT–PF2 (EMD-22456, 7JSJ) complexes. All other data are available from the corresponding authors upon reasonable request. Source data are provided with this paper.
References
Spencer, A. F. & Lowenstein, J. M. The supply of precursors for the synthesis of fatty acids. J. Biol. Chem. 237, 3640–3648 (1962).
Ruderman, N. B., Saha, A. K., Vavvas, D. & Witters, L. A. Malonyl-CoA, fuel sensing, and insulin resistance. Am. J. Physiol. Endocrinol. Metabol. 276, E1–E18 (1999).
Sul, H. S. & Smith, S. in Biochemistry of Lipids, Lipoproteins and Membranes (eds Vance, D. E. & Vance, J. E.) 155–190 (Elsevier, 2008).
Fang, M. & Lowenstein, J. M. Citrate and the conversion of carbohydrate into fat. The regulation of fatty acid synthesis by rat liver extracts. Biochem. J. 105, 803–811 (1967).
Shrago, E., Spennetta, T. & Gordon, E. Fatty acid synthesis in human adipose tissue. J. Biol. Chem. 244, 2761–2766 (1969).
Thevenon, J. et al. Mutations in SLC13A5 cause autosomal-recessive epileptic encephalopathy with seizure onset in the first days of life. Am. J. Hum. Genet. 95, 113–120 (2014).
Hardies, K. et al. Recessive mutations in SLC13A5 result in a loss of citrate transport and cause neonatal epilepsy, developmental delay and teeth hypoplasia. Brain 138, 3238–3250 (2015).
Klotz, J., Porter, B. E., Colas, C., Schlessinger, A. & Pajor, A. M. Mutations in the Na+/citrate cotransporter NaCT (SLC13A5) in pediatric patients with epilepsy and developmental delay. Mol. Med. 22, 310–321 (2016).
Wellen, K. E. et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 (2009).
Inoue, K., Zhuang, L., Maddox, D. M., Smith, S. B. & Ganapathy, V. Structure, function, and expression pattern of a novel sodium-coupled citrate transporter (NaCT) cloned from mammalian brain. J. Biol. Chem. 277, 39469–39476 (2002).
Inoue, K. et al. Functional features and genomic organization of mouse NaCT, a sodium-coupled transporter for tricarboxylic acid cycle intermediates. Biochem. J. 378, 949–957 (2004).
Gopal, E. et al. Expression and functional features of NaCT, a sodium-coupled citrate transporter, in human and rat livers and cell lines. Am. J. Physiol. Gastrointest. Liver Physiol. 292, G402–G408 (2007).
Li, L. et al. SLC13A5 is a novel transcriptional target of the pregnane X receptor and sensitizes drug-induced steatosis in human liver. Mol. Pharmacol. 87, 674–682 (2015).
Brachs, S. et al. Inhibition of citrate cotransporter Slc13a5/mINDY by RNAi improves hepatic insulin sensitivity and prevents diet-induced non-alcoholic fatty liver disease in mice. Mol. Metab. 5, 1072–1082 (2016).
Birkenfeld, A. L. et al. Deletion of the mammalian INDY homolog mimics aspects of dietary restriction and protects against adiposity and insulin resistance in mice. Cell Metab. 14, 184–195 (2011).
Rogina, B., Reenan, R. A., Nilsen, S. P. & Helfand, S. L. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science 290, 2137–2140 (2000).
Wang, P. Y. et al. Long-lived Indy and calorie restriction interact to extend life span. Proc. Natl Acad. Sci. USA 106, 9262–9267 (2009).
Fei, Y. J. et al. Relevance of NAC-2, an Na+-coupled citrate transporter, to life span, body size and fat content in Caenorhabditis elegans. Biochem. J. 379, 191–198 (2004).
Bergeron, M. J., Clémençon, B., Hediger, M. A. & Markovich, D. SLC13 family of Na+-coupled di- and tri-carboxylate/sulfate transporters. Mol. Aspects Med. 34, 299–312 (2013).
Pajor, A. M. Sodium-coupled dicarboxylate and citrate transporters from the SLC13 family. Pflugers Arch. 466, 119–130 (2014).
Joshi, A. D. & Pajor, A. M. Role of conserved prolines in the structure and function of the Na+/dicarboxylate cotransporter 1, NaDC1. Biochemistry 45, 4231–4239 (2006).
Willmes, D. M. et al. The longevity gene INDY (I’m not dead yet) in metabolic control: potential as pharmacological target. Pharmacol. Ther. 185, 1–11 (2018).
Huard, K. et al. Discovery and characterization of novel inhibitors of the sodium-coupled citrate transporter (NaCT or SLC13A5). Sci. Rep. 5, 17391 (2015).
Huard, K. et al. Optimization of a dicarboxylic series for in vivo inhibition of citrate transport by the solute carrier 13 (SLC13) family. J. Med. Chem. 59, 1165–1175 (2016).
Pajor, A. M. et al. Molecular basis for inhibition of the Na+/citrate transporter NaCT (SLC13A5) by dicarboxylate inhibitors. Mol. Pharmacol. 90, 755–765 (2016).
Rives, M. L., Shaw, M., Zhu, B., Hinke, S. A. & Wickenden, A. D. State-dependent allosteric inhibition of the human SLC13A5 citrate transporter by hydroxysuccinic acids, PF-06649298 and PF-06761281. Mol. Pharmacol. 90, 766–774 (2016).
Mancusso, R., Gregorio, G. G., Liu, Q. & Wang, D. N. Structure and mechanism of a bacterial sodium-dependent dicarboxylate transporter. Nature 491, 622–626 (2012).
Nie, R., Stark, S., Symersky, J., Kaplan, R. S. & Lu, M. Structure and function of the divalent anion/Na+ symporter from Vibrio cholerae and a humanized variant. Nat. Commun. 8, 15009 (2017).
Sauer, D. B. et al. Structural basis for the reaction cycle of DASS dicarboxylate transporters. eLife 9, e61350 (2020).
Schlessinger, A., Sun, N. N., Colas, C. & Pajor, A. M. Determinants of substrate and cation transport in the human Na+/dicarboxylate cotransporter NaDC3. J. Biol. Chem. 289, 16998–17008 (2014).
Colas, C., Pajor, A. M. & Schlessinger, A. Structure-based identification of inhibitors for the SLC13 family of Na+/dicarboxylate cotransporters. Biochemistry 54, 4900–4908 (2015).
Inoue, K., Zhuang, L., Maddox, D. M., Smith, S. B. & Ganapathy, V. Human sodium-coupled citrate transporter, the orthologue of Drosophila Indy, as a novel target for lithium action. Biochem. J. 374, 21–26 (2003).
Gopal, E. et al. Species-specific influence of lithium on the activity of SLC13A5 (NaCT): lithium-induced activation is specific for the transporter in primates. J. Pharmacol. Exp. Ther. 353, 17–26 (2015).
Mancusso, R., Karpowich, N. K., Czyzewski, B. K. & Wang, D. N. Simple screening method for improving membrane protein thermostability. Methods 55, 324–329 (2011).
Auer, M. et al. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry 40, 6628–6635 (2001).
Tan, Y. Z. et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nat. Methods 14, 793–796 (2017).
Mulligan, C. et al. The bacterial dicarboxylate transporter VcINDY uses a two-domain elevator-type mechanism. Nat. Struct. Mol. Biol. 23, 256–263 (2016).
Drew, D. & Boudker, O. Shared molecular mechanisms of membrane transporters. Annu. Rev. Biochem. 85, 543–572 (2016).
Garaeva, A. A. & Slotboom, D. J. Elevator-type mechanisms of membrane transport. Biochem. Soc. Trans. 48, 1227–1241 (2020).
Khamaysi, A., Aharon, S., Eini-Rider, H. & Ohana, E. A dynamic anchor domain in slc13 transporters controls metabolite transport. J. Biol. Chem. 295, 8155–8163 (2020).
Makwana, K. M. & Mahalakshmi, R. Implications of aromatic–aromatic interactions: from protein structures to peptide models. Protein Sci. 24, 1920–1933 (2015).
Nayal, M. & Di Cera, E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 256, 228–234 (1996).
Pajor, A. M. Conformationally sensitive residues in transmembrane domain 9 of the Na+/dicarboxylate co-transporter. J. Biol. Chem. 276, 29961–29968 (2001).
Dewulf, J. P. et al. SLC13A3 variants cause acute reversible leukoencephalopathy and α-ketoglutarate accumulation. Ann. Neurol. 85, 385–395 (2019).
Law, C. J., Yang, Q., Soudant, C., Maloney, P. C. & Wang, D. N. Kinetic evidence is consistent with the rocker-switch mechanism of membrane transport by GlpT. Biochemistry 46, 12190–12197 (2007).
Law, C. J., Enkavi, G., Wang, D. N. & Tajkhorshid, E. Structural basis of substrate selectivity in the glycerol-3-phosphate: phosphate antiporter GlpT. Biophys. J. 97, 1346–1353 (2009).
Slotboom, D. J., Duurkens, R. H., Olieman, K. & Erkens, G. B. Static light scattering to characterize membrane proteins in detergent solution. Methods 46, 73–82 (2008).
Waight, A. B., Love, J. & Wang, D. N. Structure and mechanism of a pentameric formate channel. Nat. Struct. Mol. Biol. 17, 31–37 (2010).
Kendrick, B. S., Kerwin, B. A., Chang, B. S. & Philo, J. S. Online size-exclusion high-performance liquid chromatography light scattering and differential refractometry methods to determine degree of polymer conjugation to proteins and protein–protein or protein–ligand association states. Anal. Biochem. 299, 136–146 (2001).
Huynh, K. W. et al. CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1. Nat. Commun. 9, 900 (2018).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Rice, W. J. et al. Routine determination of ice thickness for cryo-EM grids. J. Struct. Biol. 204, 38–44 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Lander, G. C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Dang, S. et al. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017).
Zivanov, J., Nakane, T. & Scheres, S. H. W. Estimation of high-order aberrations and anisotropic magnification from cryo-EM data sets in RELION-3.1. IUCrJ 7, 253–267 (2020).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Kühlbrandt, W., Wang, D. N. & Fujiyoshi, Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature 367, 614–621 (1994).
Mitsuoka, K. et al. The structure of bacteriorhodopsin at 3.0 Å resolution based on electron crystallography: implication of the charge distribution. J. Mol. Biol. 286, 861–882 (1999).
Bartesaghi, A., Matthies, D., Banerjee, S., Merk, A. & Subramaniam, S. Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy. Proc. Natl Acad. Sci. USA 111, 11709–11714 (2014).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
DeLano, W. L. The PyMOL User’s Manual (DeLano Scientific, 2002).
Acknowledgements
This work was financially supported by the NIH (R01NS108151 and R01GM121994), the G. Harold and Leila Y. Mathers Foundation, the TESS Research Foundation, the American Epilepsy Society and Pfizer. D.B.S. was supported by the American Cancer Society Postdoctoral Fellowship (129844-PF-17-135-01-TBE) and Department of Defense Horizon Award (W81XWH-16-1-0153). J.K.H. and J.A.M. were supported by the NINDS Intramural Program. We thank the following colleagues for reagents, technical assistance and discussions: Y. Cheng, N. Coudray, K. Huard, T. Kawate, J. Li, R. Mancusso, J. Marden, F. Ono and J. Zhao; the staff at the following facilities for assistance in grid screening and data collection: K. Maruthi at the Simons Electron Microscopy Center at the New York Structural Biology Center, H. Scott at the Pacific Northwest Center for Cryo-EM and K. Dancel from the NYU Microscopy Core. Electron microscopy data processing used computing resources at the HPC Facility of NYULMC, and we were assisted by A. Siavosh-Haghighi and M. Costantino.
Author information
Authors and Affiliations
Contributions
J.S. expressed and purified the protein. J.S., N.K.K. and J.K.H. conducted biochemical studies. D.B.S. froze grids. D.B.S., B.W. and W.J.R. collected and processed the cryo-EM images. D.B.S. built the atomic models. D.B.S and D.-N.W. analysed the structures. D.B.S., J.K.H., J.A.M., W.J.R. and D.-N.W. wrote the manuscript. All authors participated in the discussion and manuscript editing. D.-N.W. supervised the research.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Raimund Dutzler and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Kinetic cycle of NaCT and molecular structures of its substrates and inhibitors.
a, b, Kinetic cycle (a) and schematic model (b) of the SLC13 transport cycle. Co, outward-facing conformation; Ci, inward-facing conformation; S, substrate. The number of cotransported Na+ for different SLC13 transporters varies between 3 and 4, but only two are shown here. All available biochemical evidence indicates that sodium ions bind before and release after the substrate. c, Molecular structures of the NaCT substrate, citrate and various inhibitors. d, IC50 of inhibitors for NaCT and the dicarboxylate transporters NaDC1 and NaDC323,24,25. Whereas PF2 is highly selective towards NaCT, PF4 and PF4a inhibit all three human di-/tricarboxylate transporters.
Extended Data Fig. 2 Purification and functional characterization of human NaCT.
a, Michaelis–Menten plot showing the citrate dependence of Na+-driven radioactive citrate uptake into HEK293 cells that expressed eGFP–NaCT. HEK293 cells transfected with an eGFP vector were used as a control. All data points include six biological replicates from two independent experiments, with error bars indicating the s.d. Inset, representative confocal image of HEK293 cells transfected with the eGFP–NaCT construct, from four biological replicates. Scale bar, 10 μm. b, Analytical fluorescence SEC of detergent-solubilized cell lysate of Hi5 cells overexpressing an eGFP–NaCT construct. Peak height represents the protein concentration, whereas the peak sharpness indicates the protein homogeneity. The cell lysate was solubilized in DDM detergent, incubated with various compounds at 37 °C, and loaded onto an analytical SEC column on HPLC. c, Preparative SEC of NaCT following Ni2+-NTA affinity purification. d, Representative SDS–PAGE of purified NaCT, from twenty biological replicates. e, NaCT binding to citrate in detergent solution as measured by tryptophan fluorescence quenching. All points include three biological replicates, with error bars indicating the s.d. The Kd was found to be 148 ± 28 μM. f, Molecular mass measurements of DDM-purified NaCT using multi-angle light scattering. The measured mass of 125 ± 2 kDa agrees with the molecular weight of a dimeric NaCT of 126.124 kDa calculated from the protein sequence.
Extended Data Fig. 3 Characterization of the NaCT–citrate cryo-EM specimens and flow chart of image processing.
a, Violin plot showing the distribution of ice thickness in electron micrographs from specimens tilted at 0°, 20°, 40° and 50°. The plot widths correspond to ice thickness distribution. Theoretically, the ice thickness at 20°, 40° and 50° tilts would increase from 0° by 6%, 30% and 56%, respectively. The actual number of electron micrographs with ultra-thin ice (5–20 nm) decreased significantly with the tilt angle. b, Violin plot showing the distribution of the average horizontal particle displacements from the first five frames of each electron micrograph. The beam-induced particle displacements increased with the tilt angle. c, Violin plot showing the distribution of micrograph CTF fit resolution of the micrographs. The image quality markedly deteriorated for those recorded from 50° tilted specimens. d, Flow chart of image processing of the NaCT–citrate images. Only images collected from specimens tilted at 0°, 20° and 40° were included in the processing and the generation of the finals maps.
Extended Data Fig. 4 Cryo-EM data collection from 0°, 20° and 40° tilted specimens and image processing of the NaCT–citrate complex.
a, Orientation distribution of particles from a NaCT–citrate complex reconstruction using only particles from 0° tilt micrographs. At 0° sample tilt most of the particles are top views (viewed along the membrane normal). Side views (viewed from within the membrane plane) are relatively rare. The number of side views and top views differ by three orders of magnitude, indicating a considerable degree of preferred orientation. b, c, Orientation distribution of particles from a NaCT–citrate complex reconstruction using particles from 40° specimen tilt (b) and all micrographs at 0°, 20° and 40° specimen tilts (c). With tilting, the orientation distribution of particles becomes much more isotropic, alleviating the preferred orientation problem. d, e, The 30 most populous classes from two-dimensional classification of particles from the 0° (d) and 40° (e) tilted specimens. The 0° classes are dominated by top views, with few side and oblique views. By contrast, the 40° micrographs include clear side- and oblique-view classes.
Extended Data Fig. 5 Structure determination of NaCT–citrate complex.
a, Cryo-EM map FSC curve of the NaCT–citrate complex reconstruction using all micrographs. Arrows indicate the nominal map resolution of 3.04 Å, based on a threshold of FSC = 0.143. b, Directional FSC curves of the NaCT–citrate complex reconstruction. Each purple trace is an individual FSC calculated from a conical wedge of the overall spherical shell, sampled on a 500-point Fibonacci spherical grid. The global FSC curve (the yellow trace), as calculated by averaging all directional FSC curves, also indicates a resolution of 3.04 Å. c, Mask used for refinement using cryoSPARC. d, Local resolution of the map. e, Example cryo-EM densities showing the quality of the chain tracing of the NaCT–citrate model. All of the key helices that are involved in citrate and sodium ion binding are shown. The density for peripheral helix TM1 is poorly resolved, with the helix loosely attached to the rest of the protein. f, Model of NaCT dimer. The scaffold domain and the transport domain in each protomer is coloured green and pink, respectively. g, Model of the NaCT protomer as viewed from the cytosol. C1 symmetry was used for the image reconstruction and model refinement. The two protomers are identical, with a root mean square deviation of 0.002 Å.
Extended Data Fig. 6 Features of the NaCT–citrate structure.
a, Cryo-EM density map around the citrate-binding sites. All of the densities are shown at the same contour levels. The density for citrate is coloured red. b, Electrostatic surface of the citrate-binding site. The sodium ions at Na1 and Na2 were included in the calculations. c, Overlay of the NaCT–citrate and VcIndy–succinate (PDB: 5UL7) structures, along with their respective substrates, shown in green and grey, respectively. d, Locations of the SLC13A5-epilepsy missense mutations within the NaCT structure as viewed from the cytosol. e, Sequence alignments of the first SNT motif (left), L5ab–TM5b (centre) and second SNT motif (right) of SLC13 family proteins and bacterial homologues. The second SNT motif in NaCT has a sequence of Ser-Asn-Val. f, Interaction of Lys107 and Arg108 on H4c with other residues on H6b and TM7. g, Aromatic clusters near TM6.
Extended Data Fig. 7 Cryo-EM data collection from tilted specimens and reconstruction FSC curve of the NaCT–PF2 complex.
a, Orientation distribution of particles from a NaCT–PF2 complex reconstruction using only particles from images of 0° tilt specimens. At 0° sample tilt most of the particles are top views, whereas side views are relatively rare. The number of side views and top views differ by up to three orders of magnitude, indicating a considerable degree of preferred orientation. b, c, Orientation distribution of particles from a NaCT–PF2 complex reconstruction using particles from 40° tilt (b) and all micrographs collected at 0°, 20° and 40° tilts (c). With tilting, the particle views become much more isotropic, alleviating the preferred orientation problem. d, Cryo-EM map FSC curve of the NaCT–PF2 complex reconstruction using all micrographs. e, Cryo-EM map of the NaCT–PF2 complex with a resolution of 3.12 Å. f, Local resolution of the map. g, Example cryo-EM densities showing the quality of the chain tracing of the NaCT–PF2 model. All of the key helices that are involved in PF2 and sodium ion binding are shown.
Extended Data Fig. 8 Map and structural model of the NaCT–PF2 complex.
a, b, Structure of the NaCT–PF2 complex as viewed from the membrane plane (a) and the cytosol (b). c, d, Cryo-EM density map around the PF2-binding sites. All of the densities are shown at the same contour levels. The density for PF2 is coloured red. e, PF2-binding site as viewed from within the transport domain. f, Packing of the scaffold domain side chains around PF2. The scaffold and transport domains are coloured green and pink, respectively. Residues Leu56, Ala57, Gly409 and Ile410 are shown as spheres. g, Overlay of the NaCT–citrate and NaCT–PF2 structures in green and blue, respectively. The loops enclosing Na1 and Na2 sodium-binding sites move by around 1 Å, more tightly enclosing both sites in the NaCT–PF2 complex. h, Na+-driven citrate uptake into HEK293 cells transfected with various eGFP-tagged NaCT mutants. Each data point includes three biological replicates, with error bars indicating the s.d. NaCT(G409Q) and NaCT(I410V) mutants retained wild-type level activity and were used to measure inhibition by PF2 in Fig. 4b.
Supplementary information
Source data
Rights and permissions
About this article
Cite this article
Sauer, D.B., Song, J., Wang, B. et al. Structure and inhibition mechanism of the human citrate transporter NaCT. Nature 591, 157–161 (2021). https://doi.org/10.1038/s41586-021-03230-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03230-x
This article is cited by
-
Structural and functional implications of SLC13A3 and SLC9A6 mutations: an in silico approach to understanding intellectual disability
BMC Neurology (2023)
-
Role of sodium dependent SLC13 transporter inhibitors in various metabolic disorders
Molecular and Cellular Biochemistry (2023)
-
Structural basis for inhibition of the drug efflux pump NorA from Staphylococcus aureus
Nature Chemical Biology (2022)
-
Cryo-EM structure of human glucose transporter GLUT4
Nature Communications (2022)
-
Cryo-EM structures of recombinant human sodium-potassium pump determined in three different states
Nature Communications (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.