Abstract
Long non-coding RNAs (lncRNAs) can be important components in gene-regulatory networks1, but the exact nature and extent of their involvement in human Mendelian disease is largely unknown. Here we show that genetic ablation of a lncRNA locus on human chromosome 2 causes a severe congenital limb malformation. We identified homozygous 27–63-kilobase deletions located 300 kilobases upstream of the engrailed-1 gene (EN1) in patients with a complex limb malformation featuring mesomelic shortening, syndactyly and ventral nails (dorsal dimelia). Re-engineering of the human deletions in mice resulted in a complete loss of En1 expression in the limb and a double dorsal-limb phenotype that recapitulates the human disease phenotype. Genome-wide transcriptome analysis in the developing mouse limb revealed a four-exon-long non-coding transcript within the deleted region, which we named Maenli. Functional dissection of the Maenli locus showed that its transcriptional activity is required for limb-specific En1 activation in cis, thereby fine-tuning the gene-regulatory networks controlling dorso-ventral polarity in the developing limb bud. Its loss results in the En1-related dorsal ventral limb phenotype, a subset of the full En1-associated phenotype. Our findings demonstrate that mutations involving lncRNA loci can result in human Mendelian disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All datasets have been deposited in the Gene Expression Omnibus (GEO) database and are accessible under accession number GSE137335 (or published previously under GSE84795). As we do not have consents from patients to deposit the exome sequencing and CGH array data in an open access public repository, they are available only upon request from A.S.-F. (asuperti@unil.ch) and C.R. (carlo.rivolta@iob.ch). Data can be shared for research purposes with permission of the patient or his/her legal guardian. All the variants reported in this study have been deposited into ClinVar (NCBI) and are accessible under accession numbers SCV001450636, SCV001450638 and SCV001450639. The novel lncRNA (Maenli) transcript sequence identified and reported here has been deposited into GenBank (NCBI) and is accessible under MW294174. Previously published data used in this study include H3K4me3 ChIP–seq data from E10.5 forelimb, ChIP–seq data for CTCF, Rad21, H3K4me1 and H3K4me3 from E13.5 midbrain and ChIP–seq data for H3K27ac and H3K4me3 from E13.5 liver (GSE84793), data from the Exome Aggregation Consortium (ExAC v0.3.1; http://exac.broadinstitute.org/), the Genome Aggregation Database (gnomAD v2.1.1; https://gnomad.broadinstitute.org/), the 1000 Genomes Browser (https://www.ncbi.nlm.nih.gov/variation/tools/1000genomes/), the Exome Sequencing Project (https://evs.gs.washington.edu/EVS/), the Greater Middle East Variome Project (http://igm.ucsd.edu/gme/), and the Brazilian genomic variants database (http://abraom.ib.usp.br/). Source data are provided with this paper.
Code availability
The AutoMap tool used for homozygosity mapping and haplotype analysis of exome sequencing data is publically available at https://github.com/mquinodo/AutoMap.
References
Kopp, F. & Mendell, J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018).
Hon, C. C. et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017).
Engreitz, J. M. et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016).
Loomis, C. A. et al. The mouse Engrailed-1 gene and ventral limb patterning. Nature 382, 360–363 (1996).
Wurst, W., Auerbach, A. B. & Joyner, A. L. Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development 120, 2065–2075 (1994).
Franke, M. et al. Formation of new chromatin domains determines pathogenicity of genomic duplications. Nature 538, 265–269 (2016).
Kragesteen, B. K. et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 50, 1463–1473 (2018).
Huettl, R. E. et al. Engrailed 1 mediates correct formation of limb innervation through two distinct mechanisms. PLoS ONE 10, e0118505 (2015).
Andrey, G. et al. Characterization of hundreds of regulatory landscapes in developing limbs reveals two regimes of chromatin folding. Genome Res. 27, 223–233 (2017).
Xiang, J. F. et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 24, 513–531 (2014).
Cho, S. W. et al. Promoter of lncRNA gene PVT1 Is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412 (2018).
Isoda, T. et al. Non-coding transcription instructs chromatin folding and compartmentalization to dictate enhancer-promoter communication and T cell fate. Cell 171, 103–119 (2017).
Vian, L. et al. The energetics and physiological impact of cohesin extrusion. Cell 175, 292–294 (2018).
Kraft, K. et al. Serial genomic inversions induce tissue-specific architectural stripes, gene misexpression and congenital malformations. Nat. Cell Biol. 21, 305–310 (2019).
Wang, K. C. et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011).
Gutschner, T. & Diederichs, S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 9, 703–719 (2012).
Ishii, N. et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet. 51, 1087–1099 (2006).
Broadbent, H. M. et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 17, 806–814 (2008).
Cabianca, D. S. et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 149, 819–831 (2012).
van Dijk, M. et al. HELLP babies link a novel lincRNA to the trophoblast cell cycle. J. Clin. Invest. 122, 4003–4011 (2012).
Maass, P. G. et al. A misplaced lncRNA causes brachydactyly in humans. J. Clin. Invest. 122, 3990–4002 (2012).
Castellanos-Rubio, A. et al. A long noncoding RNA associated with susceptibility to celiac disease. Science 352, 91–95 (2016).
Royer-Bertrand, B. et al. Mutations in the heat-shock protein A9 (HSPA9) gene cause the EVEN-PLUS syndrome of congenital malformations and skeletal dysplasia. Sci. Rep. 5, 17154 (2015).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 43, 11.10.1–11.10.33 (2013).
Karczewski, K. J. et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic Acids Res. 45 (D1), D840–D845 (2017).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Naslavsky, M. S. et al. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum. Mutat. 38, 751–763 (2017).
Kraft, K. et al. Deletions, inversions, duplications: engineering of structural variants using CRISPR/Cas in mice. Cell Rep. 10, 833–839 (2015).
George, S. H. et al. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl Acad. Sci. USA 104, 4455–4460 (2007).
Artus, J. & Hadjantonakis, A. K. Generation of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Methods Mol. Biol. 693, 37–56 (2011).
Liska, F. et al. Impairment of Sox9 expression in limb buds of rats homozygous for hypodactyly mutation. Folia Biol. 56, 58–65 (2010).
Wingett, S. et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Res. 4, 1310 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Durand, N. C. et al. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Lupiáñez, D. G. et al. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161, 1012–1025 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Wolock, S. L., Lopez, R. & Klein, A. M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst. 8, 281–291 (2019).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 20, 296 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Ibrahim, D. M. et al. Distinct global shifts in genomic binding profiles of limb malformation-associated HOXD13 mutations. Genome Res. 23, 2091–2102 (2013).
Lee, T. I., Johnstone, S. E. & Young, R. A. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1, 729–748 (2006).
Paliou, C. et al. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc. Natl Acad. Sci. USA 116, 12390–12399 (2019).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–W165 (2016).
Vogel, M. J., Peric-Hupkes, D. & van Steensel, B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2, 1467–1478 (2007).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Gagnon, K. T., Li, L., Janowski, B. A. & Corey, D. R. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat. Protoc. 9, 2045–2060 (2014).
Grote, P. et al. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell 24, 206–214 (2013).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Acknowledgements
This work was supported by grants MU 880/16-1 and MU 880/14-3 from the Deutsche Forschungsgemeinschaft (DFG) to S.M., by grants from the Fondation Guillaume-Gentil (Lausanne) and the Faculté de Biologie et Medecine of the University of Lausanne to A.S.-F., and by the Swiss National Science Foundation grant no. 176097 to C.R. L.A. was supported by the FOXG1 Foundation (grant P.S.7905). We thank K. Macura, C. Franke, N. Engemann, C. Hillgardt, V. Desiniotis, N. Brieske, S. Haas and K. Kraft in Berlin for technical and/or scientific support.
Author information
Authors and Affiliations
Contributions
L.A., S.M. and A.S.-F. conceived the project. C.E.S.-M., D.R.C., L.F., C.M.L., R.A., S.R. and S.N. examined the patients and provided clinical information, images and biological samples. C.E.S.-M., S.U., L.B., C.R., S.M. and A.S.-F. interpreted the clinical and imaging data for differential diagnosis and identification of candidate genes and disease molecular aetiology. S.B., M.Q. and B.R.-B. performed the NGS studies and autozygosity mappings. F.N.-B. and S.U. provided the cytogenetic study and interpretation. S.B., B.C.-X., C.C. and R.P.G. carried out Sanger sequencing analyses for the validation of the deletion breakpoints and exonic variants in all four patients. L.A. and S.M. designed the functional experiments to determine the disease molecular mechanism with the input of P.G. L.A. generated all the transgenic mouse models (knockouts and knock-ins). L.W. performed morula aggregation. L.A. performed RNA-seq, RT–qPCRs, in-situ hybridizations, skeletal preparations and phenotype analyses. L.A. performed RACE PCR and subcellular fractionation analyses. L.A. and A.M. performed ChIP–seq and ATAC-seq with the computational analyses performed by L.A. and V.H. L.A. performed cHi-C and R.S. performed the computational analysis. W.-L.C. performed the micro-computed tomography analyses. M.I.R. and A.R. performed DamID-seq with analyses by V.H. G.C. and G.A. performed scRNA-seq and C.A.P.-M. performed the computational analysis. B.T. performed high-throughput sequencing. L.A., S.M. and A.S.-F. wrote the manuscript with input from M.S., M.I.R., S.U. and P.G. and the manuscript was approved by all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Annelien Bredenoord, Howard Chang, Moisés Mallo and Andrew Wilkie for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
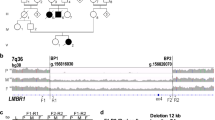
Extended Data Fig. 1 Clinical and molecular features of the affected individuals.
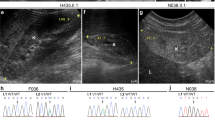
a, Photographs and X-ray of patients 1 and 4 showing the presence of ventral nails (1), normal femurs (2), severely shortened and triangular tibiae (3), triangular fibulae (4). b, Pedigrees of patients 1–4. c, Sequencing of ~3.0 and ~7.0 kb PCR fragments across the breakpoints revealing a deletion of 27,828 bp in patients 1 and 2, and a deletion of 62,789 bp with a 2,576 bp insertion at the breakpoint in patient 3, respectively. The genomic coordinates for chromosome 2 are based on the UCSC Genome Browser (GRCh38/hg38, December 2013 assembly). d, Conventional Sanger sequencing chromatograms show the homozygous mutation (c.317dupT) in patient 4. The presence of a monoallelic variant (c.317dupT) in the asymptomatic mother is also shown. The impact of the biallelic frameshift mutation (c.317dupT) on the amino acid sequence of the resulting EN1 protein is highlighted in yellow. dup, duplication; H, histidine; I, isoleucine; R, arginine; D, aspartic acid; X, stop codon.
Extended Data Fig. 2 Del(27) mouse model exhibits a limb phenotype similar to En1−/− mice.
a, Alcian blue (cartilage) and alizarin red (bone) stained limbs prepared from wild-type and Del(27) newborn mice. Fusion of digits III and IV at the level of phalanges 3 and 2 is observed for the Del(27)#H11 mutant. Digits II and III are displaced towards digit I (preaxial shift) for the Del(27)#E6 mutant. For both mutants, digits are flexed dorsally rather than ventrally. p, phalange. Scale bars, 500 μm. b, c, Ventral views (b) and high power digit views (c) of wild type and Del(27) adult fore- and hindpaws illustrating the presence of cylindrical nails (1), ventral nails formed opposite to the dorsal nails (2), normal dorsal and ectopic ventral hairs (3). The pigmented metatarsal pads (4) are elongated and hardened, resembling nails, and the fusion (5) and polydactyly of digits (6) are variable. Scale bars, 2,000 μm and 500 μm in b and c, respectively. d, μCT of wild type and Del(27) adult fore- and hindpaws show that the ventral skeletal elements, such as the falciform (green) and sesamoid bones (red), were lost or incompletely formed in Del(27) mutant paws. Scale bars, 1,000 μm. e, Normalized qRT–PCR of En1 in limbs, somites, and hindbrains prepared from wild type and Del(27) E11.5 embryos show a complete loss of mRNA expression specifically in the limb buds (P value = 1.58 × 10−4). No changes in En1 mRNA levels were detectable in the somites and hindbrains (P value = 0,012; 0,16). Data were normalized to wild type expression. We used a one-tailed t-test to evaluate the significance of En1 loss (n = 3 biologically independent wild type and mutant limb, somite, and hindbrain pairs; mean ± s.d.).
Extended Data Fig. 3 Del(27) mouse model does not exhibit sternum, ribs, or brain defects.
a, WISH of wild type and Del(27) embryos at E11.5 probed with Fgf8, Lmx1b, or Wnt7a. Limb mutants display ectopic ventral expression of the dorsalizing ge-nes Lmx1b (1) and Wnt7a (2) as well as aberrant proximoventral expansion of the Fgf8 expression domain in the AER (3). Scale bars, 1,000 μm for the whole embryos and 200 μm for the high power view of the limb buds, respectively. b, Alcian blue- (cartilage) and alizarin red- (bone) stained sternum and ribs prepared from wild type and Del(27) newborn mice show that these structures are indistinguishable between wild type and mutant mice. Scale bars, 2,000 μm. c, Intact adult brains prepared from wild type and Del(27) mice show no brain abnormalities. cb, cerebellum; ci, colliculi. Scale bars, 2,000 μm.
Extended Data Fig. 4 Maenli has two alternatively spliced isoforms.
a, Diagram of the Maenli transcripts expressed in the mouse limb bud, determined using 5′ and 3′ RACE. Ex, exon; 5′GSP1, 5′ Maenli-specific primer from exon 4; 3′GSP1, 3′ Maenli-specific primer from exon 2. b, Full-length Maenli RT–PCR analysis. RT–PCR was carried out using cDNAs generated using total RNA isolated from E9.5 forelimb bud. Agarose gel electrophoresis profile shows the presence of the expected size of DNA fragments (1016 bp and 871 bp) corresponding to Maenli-transcript 1 and Maenli-transcript 2, respectively. 1, E9.5 forelimb bud cDNAs; 2, 1 kb plus DNA ladder. c, Sequencing of the subcloned RT–PCR products illustrates the exon skipping event involving exon 3 of Maenli. The double arrowheads indicate the exon/exon junctions. d, Isoform-specific qRT–PCR of Maenli in E9.5 forelimb shows that the 4-exon Maenli transcript 1 is the major isoform (n = 3 biologically independent wild type forelimbs; mean ± s.d.). e, Normalized qRT–PCRs of En1 and Maenli in E9.5 forelimb buds prepared from wild type embryos. Maenli expression is on average 7 times lower than En1 mRNA level. Data were normalized to Maenli expression; n = 3 biologically independent wild type forelimbs; mean ± s.d. f, PhyloCSF tracks show a lack of protein coding potential for the Maenli transcript. g, Mammalian and vertebrate sequence conservation tracks show that the Maenli TSS region is highly conserved while the Maenli nucleotide sequence exhibits little conservation.
Extended Data Fig. 5 Maenli is a nuclear-enriched lncRNA.
a, Nucleotide sequence of the mouse Maenli transcript. Exons 1, 2, 3, and 4 are represented using different colours. The exon–exon junctions are highlighted in blue. The primers used for the amplification of the full Maenli cDNA are highlighted in yellow. b, Subcellular fractionation of Malat1, Tug1, Uph, En1, and Maenli. qPCR analysis of cDNAs derived from the cytoplasm, nucleoplasm, or chromatin fractions of cells obtained from E9.5 forelimb buds shows that Maenli is enriched in the nuclear fraction (75% of total RNA) (n = 3 biological replicates from 4 independent experiments; mean ± s.d.). c, ChIP–seq tracks for H3K4me3 and H3K27ac in E9.5 forelimb bud and brain show the presence of the active histone modifications at the Maenli TSS specifically in the developing limb bud, indicated with black arrowheads. No active histone modifications were detected at the Maenli TSS in the brain. d, Normalized qRT–PCRs of En1 and Maenli in E9.5, E10.5, and E11.5 forelimb buds show a decrease of Maenli expression from E9.5 to E11.5, while En1 expression is stable across the different developmental stages. Data were normalized to E11.5 En1 or Maenli expression; n = 3 biologically independent wild type forelimbs; mean ± s.d. e, ChIP–seq tracks from wild type (WT) E9.5, E10.5, and E11.5 forelimb buds (FL) show the presence of the H3K4me3 active epigenetic mark at the Maenli TSS at E9.5 but not at later stages E10.5 and E11.5. WT, wild type; FL, forelimb. f, The percentage of ectodermal Maenli-expressing cells that express Wnt7a (dorsal ectoderm marker gene), En1 (ventral ectoderm marker gene), and Fgf8 (AER marker gene) is shown. Maenli is mostly co-expressed with En1 and Fgf8 and is thus enriched in the AER and ventral ectoderm, with only a few positive cells in the dorsal ectoderm. Of these Maenli/Wnt7a positive cells, the majority also express either En1 or Fgf8.
Extended Data Fig. 6 Knock-ins and knockouts experimental design as well as quantification of Maenli expression primer design.
a, A schematic of the Maenli locus showing the primer positions used for the quantification of Maenli expression in the different alleles included in Fig. 3a and Extended Data Fig. 7c. Six pairs of qRT–PCR primers were used for the quantification of Maenli expression. Depending on the genetic manipulation, different primer pairs were used for the quantification of Maenli expression, ensuring that the same sizes of qRT–PCR products were amplified in the wild type and in the mutant samples. One pair of qRT–PCR primers (qRT–PCR-Rps9-F/R) was used as an endogenous control. The fold change (FC) was determined using the 2−ΔΔCT method. The dosage of each amplicon was normalized to the endogenous control amplicon and relative to the corresponding wild type sample. As for every genetic manipulation, the qRT–PCR results of the mutant allele were relative (normalized) to the corresponding qRT–PCR results of the wild type allele, only one wild type sample is shown in Fig. 3a and Extended Data Fig. 7c with an RQ value = 1 (100%) for simplicity. The sequences of the qRT–PCR primers used for the quantification of Maenli expression are listed in Supplementary Table 1. The exact position of the primers within the exons is indicated with a vertical dash. RE, regulatory element; Ex, exon; 3xpA, triple polyadenylation termination sequence; GFP, green fluorescent protein. b, c, e, Targeting strategy for the insertion of a triple polyadenylation termination sequence (pAS) (b), an inverted pAS (InvpAS) (c), and a GFP sequence followed by a pAS sequence (e) into exon 1 of Maenli. A schematic of the donor vector and CRISPR-mediated homologous recombination targeting strategy is shown. The primers F1, F2, R1, and R2 used for genotyping of the knock-in alleles by PCR correspond to the primers KI-PCRg-1-F, KI-PCRg-2-F, KI-PCRg-1-R, and KI-PCRg-2-R listed in Supplementary Table 1, respectively. Ex, exon; F, forward; R, reverse; GFP, green fluorescent protein. d, A schematic of the knockouts of Maenli exons and introns is shown. We note that the intron deletions, by design, do not affect the sequence of the spliced Maenli RNA. Del, deletion.
Extended Data Fig. 7 En1 activation is dependent on Maenli transcription.
a, WISH of wild type and MaenlipAS/pAS embryos at E11.5 probed with En1, Fgf8, Lmx1b, or Wnt7a. Limb mutants show an almost complete loss of En1 expression (black arrowhead), ectopic ventral expression of the dorsalizing genes Wnt7a and Lmx1b (black arrows), as well as aberrant proximoventral expansion of the Fgf8 expression domain in the AER (black arrow). d, dorsal; v, ventral. Scale bars, 200 μm. b, Poly(A)+ RNA-seq profiles of E9.5 forelimb buds show that the pAS insertion abolishes Maenli transcription in both MaenlipAS/pAS and MaenliGFP-pAS/GFP-pAS mutants. However, while MaenlipAS/pAS mutants exhibit an almost complete loss of En1 expression, MaenliGFP-pAS/GFP-pAS mutants exhibit a less severe reduction in En1 expression. A summary of the expression values for En1 and Maenli is shown. FPKM, fragments per kilobase of exon per million fragments mapped; ND, not defined; Mut, MaenlipAS/pAS. c, Normalized qRT–PCRs of Maenli and En1 in E9.5 forelimb embryos show that individual exon and intron deletions of Maenli had no significant effect on Maenli and En1 expression (P value = 0,10; 0,28; 0,14, 0,06; 0,38; 0,40; 0,04; 0,36; 0,19; 0,31; 0,04; 0,41). Data were normalized to wild type Maenli or En1 expression (n = 3 biologically independent wild type and mutant limb pairs; a one-tailed t-test was used to evaluate the significance of Maenli and En1 changes; mean ± s.d.). d, Ventral views of adult hindpaws prepared from wild type and mutant mice carrying a deletion of each of the three downstream exons and three introns of Maenli. Note that the mutant hindpaws are indistinguishable from wild type. Del, deletion. Scale bars, 2,000 μm. e, Normalized qRT–PCRs of GFP and Rps9 in E9.5 forelimb show that transcription of the orthologous GFP sequence is detected in MaenliGFP-pAS/GFP-pAS mutants but not in MaenlipAS/pAS mutants (cycle threshold undetermined). Rps9 was used as an endogenous control to assess RNA quality and quantity. Data were normalized to MaenliGFP-pAS/GFP-pAS GFP and Rps9 expression (n = 3 biologically independent mutant limb pairs; mean ± s.d.). Ex, exon; RE, regulatory element; GFP, green fluorescent protein; 3xpA, triple polyadenylation termination sequence (stop cassette); FL, forelimb.
Extended Data Fig. 8 Limb, brain, and sternum defects in En1−/− mice produced by crossings of En1+/− mice.
a, Alcian blue- (cartilage) and alizarin red- (bone) stained forelimbs prepared from wild type and En1−/− E18.5 embryos show the presence of syndactyly of the 3rd and 4th digits at the level of phalanges 3, 2, and 1 for both mutants, as well as the presence of polydactyly (VI) and fusion of digits II and VI. Note the presence of the postaxial polydactyly (VII) for En1−/−#2. p, phalange. Scale bars, 2,000 μm. b, Alcian blue- (cartilage) and alizarin red- (bone) stained hindlimbs prepared from wild type and En1−/− E18.5 embryos show the presence of ectopic ventral digits (VI) fused at the level of phalanges 1 and 3 for En1−/−#1 and En1−/−#2, respectively. p, phalange. Scale bars, 500 μm. c, Wild type and mutant sternums stained with alcian blue and alizarin red show truncated sternums (black arrows) of En1−/− mutants and abnormal placement of the ribs. The sternebrae are reduced in size and intercostal ossification is reduced and irregular (black arrowheads). Scale bars, 1,000 μm. d, Intact brains prepared from wild type and En1−/− E18.5 embryos show the absence of the colliculi in the mutants. ci, colliculi. Scale bars, 500 μm.
Extended Data Fig. 9 Maenli expression does not seem to modulate neither 3D chromatin folding nor CTCF and cohesin occupancy.
a, DamID-seq tracks from wild type mouse ES cells and in vitro culture of hindlimb buds from E11.5 embryos show that the En1 locus and its regulatory landscape are located to the nuclear interior in both cell types. mESCs, mouse embryonic stem cells; HLCCs, primary culture of cells derived from E11.5 hindlimb buds. b, Subtraction map of E9.5 MaenlipAS/pAS forelimb cHi-C signal from E9.5 wild type forelimb cHi-C signal shows no major differences between the wild type and mutant limb buds. ChIP–seq profiles for CTCF and Rad21 from wild type (WT) and MaenlipAS/pAS (Mut) E11.5 limb buds (FLHL) are shown. No major differences in CTCF or Rad21 binding were observed between the wild type and mutant limb buds, but also between the wild type limb buds and a control sample (wild type midbrain (MB)) used for the detection of limb-specific peaks. FLHL, forelimb-hindlimb; MB, midbrain; Mut, MaenlipAS/pAS; WT, wild type. The black triangle delimits the En1 TAD. The yellow segment line delimits the En1 limb regulatory domain. The black segment line delimits the end of the En1 TAD.
Extended Data Fig. 10 Maenli transcription loss affects the deposition of H3K4me3 across the En1 limb regulatory domain.
a, ATAC reads and ChIP–seq profiles for H3K27ac, H3K4me1 and H3K4me3 from wild type (WT) and MaenlipAS/pAS (Mut) E11.5 limb buds (FLHL) are shown. Major changes in chromatin accessibility and histone marks are highlighted in orange and indicated with black arrows. A control sample (liver (Li) or midbrain (MB)) used for the detection of limb-specific peaks is shown when possible. A zoom in of the En1 limb regulatory domain for H3K4me3 ChIP–seq tracks from wild type (WT), MaenlipAS/pAS (Mut) and Del(27) E11.5 limb buds (FLHL) show the loss of H3K4me3 across the En1 limb regulatory domain. Control samples including the midbrain (MB) and liver (Li) used for the detection of limb-specific peaks are shown. FLHL, forelimb-hindlimb; Li, liver; MB, midbrain; Mut, MaenlipAS/pAS; WT, wild type. The yellow segment line delimits the En1 limb regulatory domain. The black segment line delimits the end of the En1 TAD. Vertebrate conservation is measured using the Genomic Evolutionary Rate Profiling (GERP) method. b, Genetic model depicting the regulation of En1 expression in the developing limb bud. c, ChIP–seq tracks for MLL1 in E11.5 limb buds show that MLL1 is enriched at the HOTTIP/HOXA locus but not at the Maenli/En1 locus.
Supplementary information
Supplementary Information
Supplementary Note 1: Clinical descriptions of patients 1-4.
Supplementary Information
Supplementary Note 2: Nucleotide sequence of the Maenli transcript.
Supplementary Table 1
List of sgRNAs used for CRISPR targeting as well as primer sequences used in this study.
Supplementary Table 2
List of marker genes used to identify the different forelimb cell clusters in single cell RNA-sequencing data.
Rights and permissions
About this article
Cite this article
Allou, L., Balzano, S., Magg, A. et al. Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature 592, 93–98 (2021). https://doi.org/10.1038/s41586-021-03208-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03208-9
This article is cited by
-
The emerging roles of long noncoding RNAs in lymphatic vascular development and disease
Cellular and Molecular Life Sciences (2023)
-
Combined loss of CDH1 and downstream regulatory sequences drive early-onset diffuse gastric cancer and increase penetrance of hereditary diffuse gastric cancer
Gastric Cancer (2023)
-
Long non-coding RNAs: novel regulators of cellular physiology and function
Pflügers Archiv - European Journal of Physiology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.