Abstract
The repair of DNA double-strand breaks (DSBs) is essential for safeguarding genome integrity. When a DSB forms, the PI3K-related ATM kinase rapidly triggers the establishment of megabase-sized, chromatin domains decorated with phosphorylated histone H2AX (γH2AX), which act as seeds for the formation of DNA-damage response foci1. It is unclear how these foci are rapidly assembled to establish a ‘repair-prone’ environment within the nucleus. Topologically associating domains are a key feature of 3D genome organization that compartmentalize transcription and replication, but little is known about their contribution to DNA repair processes2,3. Here we show that topologically associating domains are functional units of the DNA damage response, and are instrumental for the correct establishment of γH2AX–53BP1 chromatin domains in a manner that involves one-sided cohesin-mediated loop extrusion on both sides of the DSB. We propose a model in which H2AX-containing nucleosomes are rapidly phosphorylated as they actively pass by DSB-anchored cohesin. Our work highlights the importance of chromosome conformation in the maintenance of genome integrity and demonstrates the establishment of a chromatin modification by loop extrusion.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All high-throughput sequencing data (Hi-C, ChIP–seq, 4C–seq) have been deposited to Array Express (https://www.ebi.ac.uk/arrayexpress/) under accession number E-MTAB-8851. ChIP–chip data have been deposited to Array Express under accession number E-MTAB-8793. Uncropped blots are shown in Supplementary Fig. 1. Other data (ChIP–qPCR and raw microscopy data) are available upon request.
Code availability
Source codes are available from https://github.com/LegubeDNAREPAIR/LoopExtrusion.
References
Clouaire, T., Marnef, A. & Legube, G. Taming tricky DSBs: ATM on duty. DNA Repair (Amst.) 56, 84–91 (2017).
McCord, R. P., Kaplan, N. & Giorgetti, L. Chromosome conformation capture and beyond: toward an integrative view of chromosome structure and function. Mol. Cell 77, 688–708 (2020).
Arnould, C. & Legube, G. The secret life of chromosome loops upon DNA double-strand break. J. Mol. Biol. 432, 724–736 (2020).
Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Clouaire, T. et al. Comprehensive mapping of histone modifications at DNA double-strand breaks deciphers repair pathway chromatin signatures. Mol. Cell 72, 250–262.e6 (2018).
Stewart, G. S., Wang, B., Bignell, C. R., Taylor, A. M. R. & Elledge, S. J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421, 961–966 (2003).
Caron, P. et al. Cohesin protects genes against γH2AX Induced by DNA double-strand breaks. PLoS Genet. 8, e1002460 (2012).
Natale, F. et al. Identification of the elementary structural units of the DNA damage response. Nat. Commun. 8, 15760 (2017).
Ochs, F. et al. Stabilization of chromatin topology safeguards genome integrity. Nature 574, 571–574 (2019).
Iacovoni, J. S. et al. High-resolution profiling of γH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29, 1446–1457 (2010).
Chang, L.-H., Ghosh, S. & Noordermeer, D. TADS and their borders: free movement or building a wall? J. Mol. Biol. 432, 643–652 (2020).
Caron, P. et al. Non-redundant functions of ATM and DNA-PKcs in response to DNA double-strand breaks. Cell Rep. 13, 1598–1609 (2015).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Rao, S. S. P. et al. Cohesin loss eliminates all loop domains. Cell 171, 305–320.e24 (2017).
Gelot, C. et al. The cohesin complex prevents the end joining of distant DNA double-strand ends. Mol. Cell 61, 15–26 (2016).
Meisenberg, C. et al. Repression of transcription at DNA breaks requires cohesin throughout interphase and prevents genome instability. Mol. Cell 73, 212–223.e7 (2019).
Potts, P. R., Porteus, M. H. & Yu, H. Human SMC5/6 complex promotes sister chromatid homologous recombination by recruiting the SMC1/3 cohesin complex to double-strand breaks. EMBO J. 25, 3377–3388 (2006).
Ström, L., Lindroos, H. B., Shirahige, K. & Sjögren, C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16, 1003–1015 (2004).
Unal, E. et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16, 991–1002 (2004).
Covo, S., Westmoreland, J. W., Gordenin, D. A. & Resnick, M. A. Cohesin is limiting for the suppression of DNA damage-induced recombination between homologous chromosomes. PLoS Genet. 6, e1001006 (2010).
Davidson, I. F. et al. DNA loop extrusion by human cohesin. Science 366, 1338–1345 (2019).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Rep. 15, 2038–2049 (2016).
Kim, Y., Shi, Z., Zhang, H., Finkelstein, I. J. & Yu, H. Human cohesin compacts DNA by loop extrusion. Science 366, 1345–1349 (2019).
Vian, L. et al. The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178.e20 (2018).
Schmitt, A. D. et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 17, 2042–2059 (2016).
Mirny, L. A., Imakaev, M. & Abdennur, N. Two major mechanisms of chromosome organization. Curr. Opin. Cell Biol. 58, 142–152 (2019).
Barrington, C. et al. Enhancer accessibility and CTCF occupancy underlie asymmetric TAD architecture and cell type specific genome topology. Nat. Commun. 10, 2908 (2019).
Aymard, F. et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014).
Wutz, G. et al. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36, 3573–3599 (2017).
Haarhuis, J. H. I. et al. The cohesin release factor WAPL restricts chromatin loop extension. Cell 169, 693–707.e14 (2017).
Dauban, L. et al. Regulation of cohesin-mediated chromosome folding by Eco1 and other partners. Mol. Cell 77, 1279–1293.e4 (2020).
Lee, C.-S., Lee, K., Legube, G. & Haber, J. E. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nat. Struct. Mol. Biol. 21, 103–109 (2014).
Sanders, J. T. et al. Radiation-induced DNA damage and repair effects on 3D genome organization. Nat. Commun. 11, 6178 (2020).
Kim, B.-J. et al. Genome-wide reinforcement of cohesin binding at pre-existing cohesin sites in response to ionizing radiation in human cells. J. Biol. Chem. 285, 22784–22792 (2010).
Kim, S.-T., Xu, B. & Kastan, M. B. Involvement of the cohesin protein, Smc1, in Atm-dependent and independent responses to DNA damage. Genes Dev. 16, 560–570 (2002).
Collins, P. L. et al. DNA double-strand breaks induce H2Ax phosphorylation domains in a contact-dependent manner. Nat. Commun. 11, 3158 (2020).
Li, K., Bronk, G., Kondev, J. & Haber, J. E. Yeast ATM and ATR kinases use different mechanisms to spread histone H2A phosphorylation around a DNA double-strand break. Proc. Natl Acad. Sci. USA 117, 21354–21363 (2020).
Liu, Y. et al. Very fast CRISPR on demand. Science 368, 1265–1269 (2020).
Zhang, Y. et al. The fundamental role of chromatin loop extrusion in physiological V(D)J recombination. Nature 573, 600–604 (2019).
Zhang, X. et al. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature 575, 385–389 (2019).
Gothe, H. J. et al. Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. Mol. Cell 75, 267–283.e12 (2019).
Canela, A. et al. Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol. Cell 75, 252–266.e8 (2019).
Mangeot, P. E. et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 10, 45 (2019).
Marnef, A. et al. A cohesin/HUSH- and LINC-dependent pathway controls ribosomal DNA double-strand break repair. Genes Dev. 33, 1175–1190 (2019).
Morawska, M. & Ulrich, H. D. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30, 341–351 (2013).
Matelot, M. & Noordermeer, D. Determination of high-resolution 3D chromatin organization using circular chromosome conformation capture (4C-seq). Methods Mol. Biol. 1480, 223–241 (2016).
Klein, F. A. et al. FourCSeq: analysis of 4C sequencing data. Bioinformatics 31, 3085–3091 (2015).
David, F. P. A. et al. HTSstation: a web application and open-access libraries for high-throughput sequencing data analysis. PLoS ONE 9, e85879 (2014).
Kojic, A. et al. Distinct roles of cohesin-SA1 and cohesin-SA2 in 3D chromosome organization. Nat. Struct. Mol. Biol. 25, 496–504 (2018).
Acknowledgements
We thank the genomics core facility of EMBL for high-throughput sequencing; the high-throughput sequencing core facility of the I2BC (Centre de Recherche de Gif) for facilities and expertise; F. Beckouet for advice on yeast work; J. Rispal and N. Firmin for occasional experimental help; and C. Normand for discussions. Work in the Haber laboratory was funded by grant R35 GM127029 from the US National Institutes of Health. F.Z. was supported by the National Institute of General Medical Sciences Training Grant TM32GM007122. E.R. is supported by Labex Ecofect (ANR-11-LABX-0048) of the Université de Lyon, Fondation FINOVI and by the European Research Council (ERC-StG-LS6-805500) under the European Union’s Horizon 2020 research and innovation programmes. Funding in the Legube laboratory was provided by grants from the European Research Council (ERC-2014-CoG 647344), the Agence Nationale pour la Recherche (ANR-14-CE10-0002-01 and ANR-18-CE12-0015), the Institut National Contre le Cancer (INCA), and the Ligue Nationale Contre le Cancer (LNCC). This work was supported by the Fondation pour la Recherche Médicale, grant number FDT201904007941, to C.A.
Author information
Authors and Affiliations
Contributions
C.A. performed 4C–seq, Hi-C, ChIP–seq, ChIP–chip and ChIP–qPCR experiments. A.-L.F. contributed to siRNA experiments and performed CTCF ChIP–seq. K.L. and F.Z. performed yeast strain construction and γH2A ChIP. P.C. performed ChIP–chip in SCC1 siRNA. V.R. and R.M. performed bioinformatic analyses of 4C–seq, Hi-C and ChIP–seq datasets. E.P.R. and P.E.M. provided nanoblades for CRISPR–Cas9 experiments. D.N. helped to realize and analyse 4C–seq experiments. T.C. supervised experiments in human cells and helped with library preparation. J.E.H. conceived and supervised work in yeast. G.L. conceived experiments, supervised the work and wrote the manuscript. All authors commented and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Leonid Mirny and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
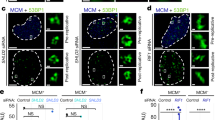
Extended Data Fig. 1 γH2AX spreads within prior TADs as revealed by 4C–seq.
a, 4C–seq tracks before DSB induction obtained for three independent biological replicates and γH2AX ChIP–seq track after DSB induction for different viewpoints (red arrows) localized at three AsiSI sites (black arrows). ChIP–seq data were smoothed using 100-kb span and 4C–seq data using a 50-kb span. b, Example of the Hi-C pattern obtained on chromosome 1 at a 500-kb resolution (left) together with a magnification at a 10-kb resolution (right). c, CTCF and calibrated-SCC1 ChIP–seq tracks. d, Average profile of CTCF ChIP–seq around all loop anchors on the genome (determined using this Hi-C dataset, Methods), validating both CTCF ChIP–seq and Hi-C datasets. e, γH2AX ChIP–seq after DSB induction. 4C–seq and CTCF ChIP–seq peak position before DSB induction are shown (peaks in blue contain a CTCF motif in the forward orientation and peaks in red a CTCF motif in the reverse orientation). f, Box plot showing γH2AX (top), 53BP1 (middle) and MDC1 (bottom) ChIP–seq quantification within the damaged TAD and neighbouring TADs for the best cleaved DSBs in DIvA cells (Methods). Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers; points, outliers (n = 153). g, γH2AX tracks around a DSB induced by CRISPR–Cas9 (top, ChIP–chip, expressed as log2[sample/input], smoothed using 100-probe windows) and by AsiSI at the same position (bottom, ChIP–seq, 50-kb smoothed). h, Top, immunofluorescence experiment showing γH2AX and DAPI staining before and after DSB induction with or without ATM inhibitor as indicated (scale bars, 10 μm). Bottom, quantification of γH2AX intensity (expressed in arbitrary units (A.U.)) in the above conditions. One representative experiment is shown (out of n = 3 biological replicates). Box plots as in f. –DSB, n = 117 nuclei; +DSB, n = 97 nuclei; +DSB + ATMi, n = 95 nuclei. i, Validation of ATR inhibitor efficiency. Western blot showing the effect of ATRi on the phosphorylation of CHK1 following treatment with hydroxyurea (HU) (n = 2). For gel source data, see Supplementary Fig. 1. j, γH2AX ChIP–seq tracks after DSB induction in untreated cells or in cells treated with an inhibitor of ATM or ATR at two DSB sites (20-kb smoothed). The differential γH2AX signal obtained after DSB induction (expressed as the log2 ratio ATMi/untreated or ATRi/untreated, grey tracks) is also shown (n = 1). k, Average profile of pATM (S1981) (left) and γH2AX (right) ChIP–seq on a 2-Mb window around the 80 best-cleaved DSBs in DIvA cells.
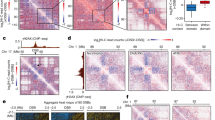
Extended Data Fig. 2 Cohesin recruitment and loop extrusion occurs at DSBs.
a, Calibrated SCC1 ChIP–seq tracks before (grey) and after (black) DSB induction (n = 1). SCC1 enrichment at DSB site is indicated by a red arrow. b, Average profile of SCC1 ChIP–seq signal centred on the 80 best-induced DSBs (left) or centred on all CTCF peaks of the genome (right) on a 10-kb window. c, Calibrated ChIP–qPCR of SCC1 in the indicated conditions at three DSB sites or a negative control region. Insets, western blots validating depletion of the proteins NIPBL (n = 1) and MRE11 (n = 2) by the corresponding siRNAs. For gel source data, see Supplementary Fig. 1. Mean ± s.e.m. for technical replicates (n = 4) of a representative experiment (out of n = 2 biological replicates). d, Averaged Hi-C matrix before (−DSB) and after DSB induction (+DSB) (observed/expected) and of the log2 ratio between damaged and undamaged cells centred on the 80 best-induced DSBs (top) or centred on eighty random TAD borders (bottom) (50-kb resolution, 5-Mb window; combined replicates). e, Averaged Hi-C contact matrix of log2[+DSB/−DSB] centred on the eighty best-induced DSBs in the two independent biological replicates. f, APA plot on a 200-kb window (10-kb resolution) before (−DSB) and after DSB induction (+DSB) in biological replicate no. 2 (replicate no. 1 shown in Fig. 2c). APAs are calculated between the DSBs and loop anchors (n = 552 pairs). The fold change between the signal (central pixel) and the background (upper left corner 5 × 5 pixels) is indicated. g, For comparison with f, APA plot on a 200-kb window (10-kb resolution) before DSB induction computed between classical loop anchors that are near DSB sites (<500 kb; n = 674 pairs for replicate 1 and n = 737 pairs for replicate 2). The fold change between the signal (central pixel) and the background (upper left corner 5 × 5 pixels) is indicated. The loop strength (quantified by the fold change between signal and background on the APA plot) is higher at loop anchors (g, replicate 1 fold-change = 5.4; replicate 2 fold-change = 5.8) than the loop strength observed at DSBs after break induction (Fig. 2c, replicate 1, fold-change = 2; f, replicate 2, fold-change = 2.3).
Extended Data Fig. 3 Loop extrusion at DSBs detected by 4C–seq.
a, 4C–seq tracks (10-kb smoothed) before and after DSB induction, obtained for three biological replicates using viewpoints localized at three DSB sites (arrows). b, 4C–seq tracks before (blue) and after (purple) DSB induction, at two DSB viewpoints. Differential 4C–seq (log2[+DSB/−DSB]) is also shown (black). c, Differential 4C–seq (log2[+DSB/−DSB]) for three viewpoints located at DSB sites and on a control region as indicated. d, Differential 4C–seq signal (log2[+DSB/−DSB]) computed on 1 Mb around four independent viewpoints located at DSBs (DSBs viewpoints, n = 11) and one control region (control viewpoint, n = 3), across four independent biological experiments (Methods). Two-sided Wilcoxon test; mean ± s.e.m. e, Western blot showing depletion of SCC1 by siRNA (n = 3). For gel source data, see Supplementary Fig. 1. f, Differential (log2) 4C–seq track in siSCC1-treated cells versus control siRNA-treated cells (in undamaged conditions) for three viewpoints. g, Genomics tracks showing 4C–seq signals before and after DSB induction in control siRNA- or siSCC1-treated cells and the differential 4C–seq signal in control siRNA- or siSCC1-treated cells (log2[+DSB/−DSB]; 10-kb smoothed). h, Average log2[+DSB/−DSB] 4C–seq, on 1 Mb around four DSB viewpoints (two biological experiments) upon treatment with control siRNA or siSCC1 (Methods) (n = 8). Two-sided Wilcoxon test. Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers; points, outliers.
Extended Data Fig. 4 ATM activity is required for loop extrusion at DSBs.
a, Hi-C maps before DSB induction of a region of chromosome 17 in control and SCC1-depleted cells. Left, 100-kb resolution; right, 25-kb resolution. b, Genomic tracks of 4C–seq before and after DSB induction in untreated or ATM-inhibitor-treated cells and of differential 4C–seq signal (log2[+DSB/−DSB] or log2[+DSB + ATMi/−DSB]; 10-kb smoothed). c, Cis interactions computed as in Extended Data Fig. 3h for four DSB viewpoints across three biological experiments, in control condition or upon ATM inhibition. Two-sided Wilcoxon test. Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers; points, outliers (n = 8).
Extended Data Fig. 5 Altered loop extrusion modifies γH2AX spreading.
a, Quantification of γH2AX intensity after DSB induction (OHT, 4 h) and upon ATM inhibition followed by different times after ATMi release (0 min, n = 172 nuclei; 5 min, n = 183 nuclei; 15 min, n = 171 nuclei; 30 min, n = 197 nuclei; 1 h, n = 189 nuclei). Treatment with OHT for 4 h without ATMi is also shown (n = 182 nuclei). One representative experiment is shown (out of n = 2 biological replicates). Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers. b, Spread of γH2AX (in bp) at the indicated time points after release from ATMi around the best cleaved DSBs (n = 71). Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers; points, outliers. c, Black, 4C–seq track before DSB induction using a DSB viewpoint. Purple, differential γH2AX signal obtained after DSB induction by ChIP–chip in SCC1-depleted versus control cells (expressed as γH2AX log2[siSCC1/siCtrl]). Light blue, differential 4C–seq signal obtained in SCC1-depleted versus control cells before DSB induction (log2[siSCC1/siCtrl]). d, Genomic tracks of γH2AX ChIP–seq signal after DSB induction in control (red) or SCC1-depleted (pink) cells and of the differential γH2AX signal obtained after DSB induction (log2[siSCC1/siCtrl], purple) at two DSB sites. e, Western blot validating the effect of the siRNA targeting WAPL on the WAPL protein level (n = 2). For gel source data, see Supplementary Fig. 1. f, Genomics tracks of γH2AX ChIP–seq after DSB induction in control or WAPL-depleted cells and of the differential γH2AX signal obtained after DSB induction (log2[siWAPL/siCtrl]) at two DSB sites and one control (no DSB) genomic locus (20-kb smoothed). g, Genomics tracks of the differential γH2A ChIP–seq signal (log2[+DSB/−DSB]) before (no IAA) or after PDS5 degradation (IAA) at two DSB sites (HO sites) in S. cerevisiae (SacCer3, coordinates in bp) (n = 1).
Extended Data Fig. 6 Increased genome-wide, DSB-induced, cohesin binding is enhanced within damaged TADs.
a, Top, contact matrix (5-kb resolution) showing log2[observed/expected] before or after DSB induction on a region showing a loop on chromosome 20 and devoid of AsiSI site (no DSB). Loops anchors are circled and indicated by red and blue bars. Bottom, genome browser screenshot showing the SCC1-calibrated ChIP–seq on the same region before and after DSB induction. Cohesin enrichment at the loop anchors (blue and red bars) is increased after DSB (black arrows) compared to before DSB (grey arrows), in agreement with increased loop strength (grey and black circles, top). b, Violin plots showing SCC1 enrichment at cohesin peaks (n = 46,194) before and after DSB induction. Paired one-sided Wilcoxon test. c, Genomic tracks of γH2AX (red) and SCC1 ChIP–seq signal before (blue) and after (purple) DSB induction. The ratio between before and after DSB induction (grey) is also shown (log2[+DSB/−DSB]; 10-kb smoothed). d, Quantification of SCC1 recruitment on loop anchors at different distances from DSB sites as indicated (from left to right, n = 1,610, 3,161, 1,930, 3,232, 4,786, 25,263, 114,461). Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum; points, outliers. e, γH2AX ChIP–seq signal and Hi-C signal at different distances from a damaged TAD on chromosome 1 before (−DSB) and after DSB induction (+DSB). Green circles, chromatin loops. f, APA plot on a 200-kb window (10-kb resolution) before (−DSB) and after DSB induction (+DSB) calculated for all loop anchors, in damaged and undamaged TADs. The fold change between the signal (central pixel) and the background (lower left corner 5 × 5 pixels) is indicated. g, Differential loop strengths in undamaged or damaged TADs (Methods), computed from Hi-C data obtained before and after DSB, from replicates 1 and 2. P values between before and after DSB are indicated (Wilcoxon test, μ = 0). The increased loop strength following DSB is significantly higher in damaged TADs than in undamaged TADs (paired two-sided Wilcoxon test) in both Hi-C replicate experiments. Replicate 1: undamaged, n = 2,936; damaged, n = 264. Replicate 2: undamaged, n = 3,181; damaged, n = 302. Box plots as in d.
Extended Data Fig. 7 DSB-induced phosphorylation of cohesin occurs in damaged TADs.
a, Genomic tracks showing γH2AX, pSMC3 S1083 and pSMC1 S966 ChIP–chip signals expressed as log2[sample/input] after DSB induction. Two damaged genomic locations are shown. b, Average profile of pSMC3 S1083 (expressed as log2([+DSB/−DSB] ChIP–seq signal) around the 80 best-induced DSBs on a 4-Mb window. c, Quantification of pSMC3 S1083 signal on loop anchors in damaged or undamaged TADs. P values between before and after DSB are indicated (paired two-sided Wilcoxon test). The increased pSMC3 S1083 enrichment on loop anchors following DSB is significantly higher in damaged TADs than in undamaged TADs (two-sided Wilcoxon test). Undamaged, n = 9,040; damaged, n = 1,626. Centre line, median; box limits, first and third quartiles; whiskers, maximum and minimum without outliers; points, outliers.
Supplementary information
Supplementary Figure
Supplementary Figure 1: Uncropped versions of the cropped Western blots presented in Extended Data Fig. 1i, 2c, 3e and 5e, with size marker indications. For each uncropped scan, the figure to which they refer is indicated above the scan. The red squares indicate which crop was used in the Extended Data figures.
Supplementary Table
Supplementary Table 1: Peak calling results (bed file) of the CTCF ChIP-seq performed before DSB induction in the DIvA cells and using MACS2. The MACS2 output “summits” is provided.
Supplementary Table
Supplementary Table 2: Peak calling results (bed file) of the SCC1 ChIP-seq performed before DSB induction in the DIvA cells and using MACS2. The MACS2 output “summits” is provided.
Rights and permissions
About this article
Cite this article
Arnould, C., Rocher, V., Finoux, AL. et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 590, 660–665 (2021). https://doi.org/10.1038/s41586-021-03193-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03193-z
This article is cited by
-
Computational methods for analysing multiscale 3D genome organization
Nature Reviews Genetics (2024)
-
A graph neural network-based interpretable framework reveals a novel DNA fragility–associated chromatin structural unit
Genome Biology (2023)
-
Chromatin compartmentalization regulates the response to DNA damage
Nature (2023)
-
Airway epithelial cGAS inhibits LPS-induced acute lung injury through CREB signaling
Cell Death & Disease (2023)
-
Determining chromatin architecture with Micro Capture-C
Nature Protocols (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.