Abstract
Lysosomes have fundamental physiological roles and have previously been implicated in Parkinson’s disease1,2,3,4,5. However, how extracellular growth factors communicate with intracellular organelles to control lysosomal function is not well understood. Here we report a lysosomal K+ channel complex that is activated by growth factors and gated by protein kinase B (AKT) that we term lysoKGF. LysoKGF consists of a pore-forming protein TMEM175 and AKT: TMEM175 is opened by conformational changes in, but not the catalytic activity of, AKT. The minor allele at rs34311866, a common variant in TMEM175, is associated with an increased risk of developing Parkinson’s disease and reduces channel currents. Reduction in lysoKGF function predisposes neurons to stress-induced damage and accelerates the accumulation of pathological α-synuclein. By contrast, the minor allele at rs3488217—another common variant of TMEM175, which is associated with a decreased risk of developing Parkinson’s disease—produces a gain-of-function in lysoKGF during cell starvation, and enables neuronal resistance to damage. Deficiency in TMEM175 leads to a loss of dopaminergic neurons and impairment in motor function in mice, and a TMEM175 loss-of-function variant is nominally associated with accelerated rates of cognitive and motor decline in humans with Parkinson’s disease. Together, our studies uncover a pathway by which extracellular growth factors regulate intracellular organelle function, and establish a targetable mechanism by which common variants of TMEM175 confer risk for Parkinson’s disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Gel pictures used to generate the immunoblot data and PCR results are available in Supplementary Fig. 1. Raw electrophysiological recording data used to generate the current-voltage relationship curves and all other data are available from the corresponding authors upon request. Websites with related data are as follows: PPMI database, www.ppmi-info.org/data; Genome Aggregation Database (gnomAD), https://gnomad.broadinstitute.org/; PD Gene database, http://www.pdgene.org/; and PDB 6WCA, https://www.rcsb.org/structure/6WCA. Source data are provided with this paper.
Change history
31 March 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-03438-x
References
Ballabio, A. & Bonifacino, J. S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118 (2020).
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012).
Wong, Y. C., Kim, S., Peng, W. & Krainc, D. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 29, 500–513 (2019).
Lie, P. P. Y. & Nixon, R. A. Lysosome trafficking and signaling in health and neurodegenerative diseases. Neurobiol. Dis. 122, 94–105 (2019).
Abeliovich, A. & Gitler, A. D. Defects in trafficking bridge Parkinson’s disease pathology and genetics. Nature 539, 207–216 (2016).
Hille, B. Ion Channels of Excitable Membranes, 3rd Edn (Sinauer Associates, 2001).
Jan, L. Y. & Jan, Y. N. Voltage-gated potassium channels and the diversity of electrical signalling. J. Physiol. 590, 2591–2599 (2012).
Xu, H. & Ren, D. Lysosomal physiology. Annu. Rev. Physiol. 77, 57–80 (2015).
Cang, C., Bekele, B. & Ren, D. The voltage-gated sodium channel TPC1 confers endolysosomal excitability. Nat. Chem. Biol. 10, 463–469 (2014).
Cang, C. et al. mTOR regulates lysosomal ATP-sensitive two-pore Na+ channels to adapt to metabolic state. Cell 152, 778–790 (2013).
Brewer, G. J., Torricelli, J. R., Evege, E. K. & Price, P. J. Optimized survival of hippocampal neurons in B27-supplemented neurobasal, a new serum-free medium combination. J. Neurosci. Res. 35, 567–576 (1993).
Cang, C., Aranda, K., Seo, Y. J., Gasnier, B. & Ren, D. TMEM175 is an organelle K+ channel regulating lysosomal function. Cell 162, 1101–1112 (2015).
Chapel, A. et al. An extended proteome map of the lysosomal membrane reveals novel potential transporters. Mol. Cell Proteomics 12, 1572–1588 (2013).
Cao, Q. et al. BK channels alleviate lysosomal storage diseases by providing positive feedback regulation of lysosomal Ca2+ release. Dev. Cell 33, 427–441 (2015).
El-Shewy, H. M., Lee, M. H., Obeid, L. M., Jaffa, A. A. & Luttrell, L. M. The insulin-like growth factor type 1 and insulin-like growth factor type 2/mannose-6-phosphate receptors independently regulate ERK1/2 activity in HEK293 cells. J. Biol. Chem. 282, 26150–26157 (2007).
Anderson, E. M. et al. BDNF–TrkB controls cocaine-induced dendritic spines in rodent nucleus accumbens dissociated from increases in addictive behaviors. Proc. Natl Acad. Sci. USA 114, 9469–9474 (2017).
Perreault, M. et al. Activation of TrkB with TAM-163 results in opposite effects on body weight in rodents and non-human primates. PLoS ONE 8, e62616 (2013).
Manning, B. D. & Toker, A. AKT/PKB signaling: navigating the network. Cell 169, 381–405 (2017).
Jo, H. et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc. Natl Acad. Sci. USA 109, 10581–10586 (2012).
Arias, E. et al. Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell 59, 270–284 (2015).
Menon, S. et al. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156, 771–785 (2014).
Oh, S., Paknejad, N. & Hite, R. K. Gating and selectivity mechanisms for the lysosomal K+ channel TMEM175. eLife 9, e53430 (2020).
Ebner, M., Lucic, I., Leonard, T. A. & Yudushkin, I. PI(3,4,5)P3 engagement restricts Akt activity to cellular membranes. Mol. Cell 65, 416–431 (2017).
Blauwendraat, C. et al. Parkinson’s disease age at onset genome-wide association study: defining heritability, genetic loci, and α-synuclein mechanisms. Mov. Disord. 34, 866–875 (2019).
Nalls, M. A. et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 46, 989–993 (2014).
Krohn, L. et al. Genetic, structural and functional evidence link TMEM175 to synucleinopathies. Ann. Neurol. 87, 139–153 (2020).
Iwaki, H. et al. Genetic risk of Parkinson disease and progression: an analysis of 13 longitudinal cohorts. Neurol. Genet. 5, e348 (2019).
Lill, C. M. et al. Impact of Parkinson’s disease risk loci on age at onset. Mov. Disord. 30, 847–850 (2015).
Davis, A. A. et al. Variants in GBA, SNCA, and MAPT influence Parkinson disease risk, age at onset, and progression. Neurobiol Aging 37, 209.e1–209.e7 (2016).
Jinn, S. et al. Functionalization of the TMEM175 p.M393T variant as a risk factor for Parkinson disease. Hum. Mol. Genet. 28, 3244–3254 (2019).
Lee, C. et al. The lysosomal potassium channel TMEM175 adopts a novel tetrameric architecture. Nature 547, 472–475 (2017).
Brunner, J. D. et al. Structural basis for ion selectivity in TMEM175 K+ channels. eLife 9, e53683 (2020).
Steinberg, B. E. et al. A cation counterflux supports lysosomal acidification. J. Cell Biol. 189, 1171–1186 (2010).
Jinn, S. et al. TMEM175 deficiency impairs lysosomal and mitochondrial function and increases α-synuclein aggregation. Proc. Natl Acad. Sci. USA 114, 2389–2394 (2017).
Aleyasin, H. et al. DJ-1 protects the nigrostriatal axis from the neurotoxin MPTP by modulation of the AKT pathway. Proc. Natl Acad. Sci. USA 107, 3186–3191 (2010).
Humbert, S. et al. The IGF-1/Akt pathway is neuroprotective in Huntington’s disease and involves Huntingtin phosphorylation by Akt. Dev. Cell 2, 831–837 (2002).
Niu, J. et al. 2′,3′-Dideoxycytidine protects dopaminergic neurons in a mouse model of Parkinson’s disease. Neurochem. Res. 42, 2996–3004 (2017).
Sakamoto, S., Miyara, M., Sanoh, S., Ohta, S. & Kotake, Y. Mild MPP+ exposure-induced glucose starvation enhances autophagosome synthesis and impairs its degradation. Sci. Rep. 7, 46668 (2017).
Whittemore, E. R., Loo, D. T., Watt, J. A. & Cotman, C. W. A detailed analysis of hydrogen peroxide-induced cell death in primary neuronal culture. Neuroscience 67, 921–932 (1995).
Goedert, M., Spillantini, M. G., Del Tredici, K. & Braak, H. 100 years of Lewy pathology. Nat. Rev. Neurol. 9, 13–24 (2013).
Fujiwara, H. et al. α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 (2002).
Luk, K. C. et al. Pathological α-synuclein transmission initiates Parkinson-like neurodegeneration in nontransgenic mice. Science 338, 949–953 (2012).
Volpicelli-Daley, L. A. et al. Exogenous α-synuclein fibrils induce Lewy body pathology leading to synaptic dysfunction and neuron death. Neuron 72, 57–71 (2011).
Gao, J., Perera, G., Bhadbhade, M., Halliday, G. M. & Dzamko, N. Autophagy activation promotes clearance of α-synuclein inclusions in fibril-seeded human neural cells. J. Biol. Chem. 294, 14241–14256 (2019).
Karpowicz, R. J. Jr et al. Selective imaging of internalized proteopathic α-synuclein seeds in primary neurons reveals mechanistic insight into transmission of synucleinopathies. J. Biol. Chem. 292, 13482–13497 (2017).
Davis, A. A., Leyns, C. E. G. & Holtzman, D. M. Intercellular spread of protein aggregates in neurodegenerative disease. Annu. Rev. Cell Dev. Biol. 34, 545–568 (2018).
Aits, S. et al. Sensitive detection of lysosomal membrane permeabilization by lysosomal galectin puncta assay. Autophagy 11, 1408–1424 (2015).
Tropea, T. F. et al. TMEM106B effect on cognition in Parkinson disease and frontotemporal dementia. Ann. Neurol. 85, 801–811 (2019).
Thomas, B. et al. Resistance to MPTP-neurotoxicity in α-synuclein knockout mice is complemented by human α-synuclein and associated with increased β-synuclein and Akt activation. PLoS ONE 6, e16706 (2011).
Datta, S. R., Brunet, A. & Greenberg, M. E. Cellular survival: a play in three Akts. Genes Dev. 13, 2905–2927 (1999).
Yang, H., Wang, H. & Jaenisch, R. Generating genetically modified mice using CRISPR/Cas-mediated genome engineering. Nat. Protoc. 9, 1956–1968 (2014).
Lu, B. et al. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell 129, 371–383 (2007).
Lu, B. et al. Extracellular calcium controls background current and neuronal excitability via an UNC79–UNC80–NALCN cation channel complex. Neuron 68, 488–499 (2010).
Bertl, A. et al. Electrical measurements on endomembranes. Science 258, 873–874 (1992).
Kao, L. R. & Megraw, T. L. RNAi in cultured Drosophila cells. Methods Mol. Biol. 247, 443–457 (2004).
Clemens, J. C. et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl Acad. Sci. USA 97, 6499–6503 (2000).
Gerdts, J., Sasaki, Y., Vohra, B., Marasa, J. & Milbrandt, J. Image-based screening identifies novel roles for IκB kinase and glycogen synthase kinase 3 in axonal degeneration. J. Biol. Chem. 286, 28011–28018 (2011).
Sasaki, Y., Vohra, B. P., Lund, F. E. & Milbrandt, J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci. 29, 5525–5535 (2009).
Wiemerslage, L. & Lee, D. Quantification of mitochondrial morphology in neurites of dopaminergic neurons using multiple parameters. J. Neurosci. Methods 262, 56–65 (2016).
Luk, K. C. et al. Molecular and biological compatibility with host alpha-synuclein influences fibril pathogenicity. Cell Rep. 16, 3373–3387 (2016).
Volpicelli-Daley, L. A., Luk, K. C. & Lee, V. M. Addition of exogenous α-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous α-synuclein to Lewy body and Lewy neurite-like aggregates. Nat. Protoc. 9, 2135–2146 (2014).
Waxman, E. A. & Giasson, B. I. Specificity and regulation of casein kinase-mediated phosphorylation of α-synuclein. J. Neuropathol. Exp. Neurol. 67, 402–416 (2008).
Angelim, M. K. S. C. et al. Embryonic macrophages and microglia ablation alter the development of dorsal root ganglion sensory neurons in mouse embryos. Glia 66, 2470–2486 (2018).
Flavin, W. P. et al. Endocytic vesicle rupture is a conserved mechanism of cellular invasion by amyloid proteins. Acta Neuropathol. 134, 629–653 (2017).
Renier, N. et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014).
Song, H. et al. Targeting Gpr52 lowers mutant HTT levels and rescues Huntington’s disease-associated phenotypes. Brain 141, 1782–1798 (2018).
Li, Z. et al. Allele-selective lowering of mutant HTT protein by HTT–LC3 linker compounds. Nature 575, 203–209 (2019).
Khaing, Z. Z. et al. Assessing forelimb function after unilateral cervical spinal cord injury: novel forelimb tasks predict lesion severity and recovery. J. Neurotrauma 29, 488–498 (2012).
Anderson, K. D., Abdul, M. & Steward, O. Quantitative assessment of deficits and recovery of forelimb motor function after cervical spinal cord injury in mice. Exp. Neurol. 190, 184–191 (2004).
Kawashita, E. et al. Altered behavior in mice with deletion of the alpha2-antiplasmin gene. PLoS ONE 9, e97947 (2014).
Gonçalves, F. M. et al. Glutamatergic system and mTOR-signaling pathway participate in the antidepressant-like effect of inosine in the tail suspension test. J. Neural Transm. (Vienna) 124, 1227–1237 (2017).
Matsuura, K., Kabuto, H., Makino, H. & Ogawa, N. Pole test is a useful method for evaluating the mouse movement disorder caused by striatal dopamine depletion. J. Neurosci. Methods 73, 45–48 (1997).
Toledo, J. B. et al. A platform for discovery: the University of Pennsylvania integrated neurodegenerative disease biobank. Alzheimers Dement. 10, 477–484 (2014).
Lucas, J. A. et al. Normative data for the Mattis Dementia Rating Scale. J. Clin. Exp. Neuropsychol. 20, 536–547 (1998).
Pigott, K. et al. Longitudinal study of normal cognition in Parkinson disease. Neurology 85, 1276–1282 (2015).
Van Deerlin, V. M. et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat. Genet. 42, 234–239 (2010).
Mata, I. F. et al. GBA variants are associated with a distinct pattern of cognitive deficits in Parkinson’s disease. Mov. Disord. 31, 95–102 (2016).
Laird, N. M. & Ware, J. H. Random-effects models for longitudinal data. Biometrics 38, 963–974 (1982).
Lee, R. S. et al. Relative expression levels rather than specific activity plays the major role in determining in vivo AKT isoform substrate specificity. Enzyme Res. 2011, 720985 (2011).
Chu, N. et al. Akt kinase activation mechanisms revealed using protein semisynthesis. Cell 174, 897–907 (2018).
Franke, T. F. et al. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell 81, 727–736 (1995).
Carpten, J. D. et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 (2007).
Parikh, C. et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc. Natl Acad. Sci. USA 109, 19368–19373 (2012).
Langston, J. W. The MPTP story. J. Parkinsons Dis. 7, S11–S19 (2017).
Choi, W. S., Kruse, S. E., Palmiter, R. D. & Xia, Z. Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc. Natl Acad. Sci. USA 105, 15136–15141 (2008).
Sanchez-Ramos, J. R., Michel, P., Weiner, W. J. & Hefti, F. Selective destruction of cultured dopaminergic neurons from fetal rat mesencephalon by 1-methyl-4-phenylpyridinium: cytochemical and morphological evidence. J. Neurochem. 50, 1934–1944 (1988).
Cohen, E. & Dillin, A. The insulin paradox: aging, proteotoxicity and neurodegeneration. Nat. Rev. Neurosci. 9, 759–767 (2008).
Acknowledgements
The work was supported in part by NIH grants 1 R01 GM133172 and 1 R01 HL147379 (to D.R.), NS088322 (to K.C.L.), and R01 NS115139, P50 NS053488 and U19-AG062418 (to A.S.C.-P.). A.S.C.-P. is also supported by the Parker Family Chair. B.L. is supported in part by the National Natural Science Foundation of China (81925012) and the Newton Advanced Fellowship (NAF_R1_191045). We thank the Transgenic and Chimeric Mouse Core of the University of Pennsylvania for the generation of all transgenic lines (supported by NIH centre grants P30DK050306, P30DK019525, and P30CA016520); A. Caputo for help with preparing the preformed α-syn fibrils; T. O’Brien for discussion on behavioural studies; and D. Weintraub, V. Van Deerlin and J. Trojanowski for help with human cohort studies. Some data used in the preparation of this Article were obtained from the PPMI database (www.ppmi-info.org/data). PPMI—a public–private partnership—is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, including Abbvie, Allergan, Amathus therapeutics, Avid Radiopharmaceuticals, Biogen, BioLegend, Bristol–Myers Squibb, Celgene, Denali, GE Healthcare, Genentech, GlaxoSmithKline, Golub Capital, Handl Therapeutics, Insitro, Janssen Neuroscience, Lilly, Lundbeck, Merck, Meso Scale Discovery, Pfizer, Piramal, Prevail Therapeutics, Roche, Sanofi Genzyme, Servier, Takeda, Teva, UCB, Verily and Voyager Therapeutics. Up-to-date information on the study is available at www.ppmi-info.org.
Author information
Authors and Affiliations
Contributions
J.W. performed all the electrophysiology studies, the neuronal damage analysis, the α-syn PFF seeding assay, galectin 3 assay, BODIPY–pepstatin A assay, autophagosome fusion assay and pH imaging in Fig. 5a and Extended Data Figs. 8–10. Z.L. and J.W. carried out the protein chemistry, RT–PCR analysis and S2 cell Akt knockdown studies. K.A., J.L. and D.R. developed cDNA constructs and mouse lines. C.C. performed earlier exploratory recordings that demonstrated that the TMEM175-knockout lysosomes lack TMEM175 currents and that SC79 activates TMEM175. Y.L. and K.C.L. performed TH immunostaining comparing the wild-type and homozygous mouse brains, and prepared and validated the α-syn PFFs. L.Y., H.W. and J.Y. performed the whole-tissue immunolabelling, optical clearing and light-sheet 3D imaging analysis comparing the wild-type and heterozygous mouse brains. H.S. and B.L. performed the mouse behavioural studies. T.F.T. and A.S.C.-P. carried out the clinical studies. D.R., J.W., B.L., J.Y., A.S.C.-P. and K.C.L. designed experiments and wrote the manuscript with contributions from all the authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks David Stone and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Lysosomal currents activated by growth factors.
Lysosomal currents were recorded using a ramp protocol (−100 mV to 100 mV ramp in 1 s with holding voltage of 0 mV) as illustrated in Fig. 1. a, Sizes of K+ currents (IK) recorded at varying voltages (Ψ, −100 mV to 100 mV) from midbrain neurons with (stv) or without (fed) overnight starvation in DMEM containing no B27 nutrient supplement. Averaged IK sizes (at 100 mV, recorded with 150 mM K+-containing bath) are in the right bar graph. Data are mean ± s.e.m. Numbers of recordings are in parentheses. *P < 0.0001, unpaired two-tailed t-test. b, Na+ currents recorded from hippocampal neurons with and without starvation demonstrating that B27 does not activate Na+ currents. The averaged current sizes (at −100 mV) are in the right bar graph (P = 0.226, unpaired two-tailed t-test). Solutions used in the recordings were the same as those used to record the IK in Fig. 1a, b, except that K+ in the bath was replaced with Na+ and 1 μM PI(3,5)P2 was added in the bath (lysosomal Na+ channel requires PI(3,5)P2 for maximum activation). c, IK recorded from TMEM175-transfected HEK293T cells starved in HBSS followed by refeeding with NGF (100 ng ml−1 for 3 h) or BDNF (10 ng ml−1 for 3 h), demonstrating that BDNF and NGF do not activate TMEM175 in HEK293T cells. Activation by insulin (right) was used as a positive control for receptor activation (see Fig. 1f for insulin). Averaged IK sizes (recorded with K+-containing bath, at 100 mV) are in the right bar graph. d, IK recorded from B27-replete midbrain and cortical neurons cultured from wild-type, heterozygous or TMEM175-knockout mice. Averaged IK sizes (at 100 mV, recorded with 150 mM K+-containing bath) are in the right bar graph. Data are mean ± s.e.m. Numbers of recordings are in parentheses. *P < 0.001, unpaired two-tailed t-test. P values (compared to wild type) are as follows: midbrain, P = 0.0007 for heterozygous, P < 0.0001 for knockout; cortex, P < 0.0001 for knockout. e, IK recorded from wild-type and TMEM175-knockout midbrain neurons before (fed) or after (stv) starvation (overnight in DMEM). An AKT activator SC79 (10 μM) was applied to the recording bath during some of the recordings (blue and red traces). Bar graphs in the right show averaged IK sizes (at 100 mV). Data are mean ± s.e.m. Numbers of recordings are in parentheses. Arrows are used to indicate curves that overlap and are not easily distinguished.
Extended Data Fig. 2 AKT is required for the reconstitution of human TMEM175 as a functional ion channel.
a–f, Whole-cell currents were recorded from Drosophila S2 cells without (a–c, h) or with (d–f) endogenous Akt (‘dAKT’) knocked down using dsRNA treatment. a, d, Nontransfected S2 cells. b, e, h, S2 cells transfected with human TMEM175 alone. c, f, S2 cells cotransfected with human TMEM175 and human AKT1. A western blot showing the reduction of Drosophila Akt protein by dsRNA treatment is shown in the inset in d. For gel source data, see Supplementary Fig. 1. g, Summary of current amplitudes (at −100 mV, recorded in K+-containing bath with or without SC79). Recordings were done using a ramp protocol (−100 mV to 100 mV in 1 s, Vh = 0 mV). Bath solution contained 150 mM K+, 150 mM K+ with SC79 (an activator of human AKT) or 150 mM NMDG with SC79, as indicated. In a, b, d, e, arrows are used to indicate curves that overlap and are not easily distinguished. Black, recorded from bath containing K+; blue, recorded from bath containing K+ and SC79; red, recorded from bath containing NMDG and SC79. Data are mean ± s.e.m. Numbers of recordings are in parentheses. In g, *P ≤ 0.05 (compared with those from cells cotransfected with human TMEM175 and human AKT). P values (unpaired two-tailed t-tests) are as follows: without dsRNA treatment, P < 0.0001 for TMEM175 vs nontransfected, TMEM175+AKT vs nontransfected and TMEM175+AKT vs TMEM175; with dsRNA treatment, P = 0.1253 for TMEM175 vs nontransfected, P < 0.0001 for TMEM175+AKT vs TMEM175 and TMEM175+AKT vs nontransfected. h, Whole-cell currents recorded from S2 cells transfected with human TMEM175 alone, with (middle) or without (left) recombinant human AKT1 protein (1 μg ml−1) included in the pipette solution. Right, summary of current amplitudes (at −100 mV; b and g show an additional control without AKT protein in pipette. We further tested the necessary and sufficient role for AKT in ITMEM175 by reconstituting TMEM175 in the Drosophila S2 cell line, which does not have endogenous TMEM175. In HEK293T cells, cotransfection of AKT1 did not further increase ITMEM175 (Extended Data Fig. 6a), consistent with the idea that the three mammalian AKTs are already abundantly expressed in the cells79. S2 cells do not have TMEM175 (which is absent in insects), and have only one Akt gene, making it easier to knock down. In addition, the Drosophila Akt (dAKT) protein is not well-conserved with the mammalian AKTs (61% identity to human AKT1) and is less likely to support human TMEM175 function. For technical reasons, we were unable to enlarge S2 cell lysosomes suitable for patch-clamp recording. We therefore recorded whole-cell currents as an assay for functional channels. Transfecting human TMEM175 alone into S2 cells generated little current (about −20 pA at −100 mV) above the level of endogenous K+ currents (a, b, g). In addition, knocking down Drosophila Akt with dsRNA55 completely eliminated the small ITMEM175 (d, e, g). Cotransfecting human TMEM175 with a human AKT1 (hAKT1), however, led to robust ITMEM175, either with or without the endogenous Drosophila Akt knocked down (c, f, g). The currents were potentiated by SC79 in human AKT cotransfected cells but not in those transfected with TMEM175 alone, supporting the notion that SC79 acts on mammalian AKTs to open the channel. We finally tested whether purified recombinant AKT protein is sufficient to activate TMEM175. In S2 cells transfected with human TMEM175 alone (which had minimal current), application of SC79 into the bath increased the currents to −417.0 ± 53.9 pA (h) (n = 3, −100 mV) when human AKT1 protein was introduced into cytosol via pipette solution dialysis. Together, our data suggest that AKT is obligatory for a functional mammalian TMEM175 channel.
Extended Data Fig. 3 mTOR is not required for the activation of TMEM175 by AKT.
a–c, Lysosomal IK currents were recorded from TMEM175-tranfected HEK293T cells with mTOR (a component of both mTORC1 and mTORC2 complexes) (a), Raptor (an mTORC1 component) (b) or Rictor (an mTORC2 component) (c) knocked down with short hairpin (sh)RNA lentivirus10. d, Lysosomal IK recorded from TMEM175-tranfected HEK293T cells treated with rapamycin (2 μM, 1 h). Figure 1f provides comparisons with those with no knockdown or rapamycin treatment. *P ≤ 0.05. P values (unpaired two-tailed t-tests; K vs K+SC79 groups) are: P < 0.0001 (mTor shRNA), P = 0.0013 (Raptor shRNA), P < 0.0001 (Rictor shRNA). e–h, Control for mTOR knockdown. Lysosomal Na+ currents were recorded from HEK293T cells transfected with TPC2 as positive controls for mTOR knockdown. TPC2 is a lysosomal Na+ channel that is inhibited by ATP. The ATP sensitivity requires mTORC1 but not mTORC2; knocking down mTOR or Raptor—but not Rictor—reduces the ATP sensitivity10. In f, arrows are used to indicate traces that overlap and are not easily distinguished. Black, recorded before 1 mM ATP was added to the bath; red, recorded after 1 mM ATP was added to the bath. Data are mean ± s.e.m. Numbers of recordings are in parentheses.
Extended Data Fig. 4 Characterization of TMEM175–AKT interaction and its role in TMEM175 activation.
a, Association between heterologously expressed TMEM175 and AKT. HEK293T cells were transfected with GFP-tagged wild-type or mutant TMEM175 and HA-tagged human AKT1 (HA–AKT), as indicated. Lysates were immunoprecipitated with anti-GFP and blotted with anti-GFP or with anti-AKT. Whole-cell lysate was also blotted for input control. b–d, Experiments demonstrating that S241 is not essential for TMEM175 activation. b, Sequence alignment of the region around S241 of TMEM175. GenBank access numbers are in parentheses. c, Lysosomal IK recorded from HEK293T cells transfected with S241A/T338A or S241A substitutions. Averaged IK sizes (at 100 mV) are shown in the bar graphs. Data are mean ± s.e.m. Numbers of lysosomes recorded are in parentheses. d, Association between wild-type or S241A-mutant TMEM175 and AKT. HA-tagged AKT was transfected alone (lane 1) in HEK293T cells or together with GFP-tagged wild-type or S241A-mutant human TMEM175 (lanes 2 and 3). Whole-cell lysates were blotted with anti-GFP (bottom) or anti-HA (middle), or immunoprecipitated with anti-GFP followed by immunoblotting with anti-HA (top). In c, arrows are used to indicate curves that overlap and are not easily distinguished. Black, recorded from bath containing K+; blue, recorded from bath containing K+ and SC79. e, Mapping the AKT-interacting region to the second repeat of TMEM175. A diagram illustrating the GFP-tagged TMEM175 fragments used in the experiments is shown. Lysates from HEK293T cells transfected with HA-tagged AKT1 (HA–AKT) alone (lane 1) or together with GFP-tagged TMEM175 fragments (lanes 2–6) were blotted with anti-GFP (bottom) or anti-HA (middle) or immunoprecipitated with anti-GFP followed by blotting with anti-HA (top). f, Phosphorylation status of T308 and S473 of TMEM175-associated AKT. HEK293T cells were transfected with GFP-tagged TMEM175 (with nontransfected cells as control). Endogenous AKT associated with TMEM175 was pulled down with immunoprecipitation using anti-GFP and was probed with antibodies against total AKT (pan-AKT), AKT phosphorylated at T308 (pAKT(T308)) or at S473 (pAKT(S473)). Total cell lysates (input) before immunoprecipitation were also loaded for comparison. g, Association between TMEM175(M393T) and TMEM175(Q65P) and AKT. Lysates from HEK293T cells transfected with GFP-tagged wild-type or mutant TMEM175 (with nontransfected cells as control) were blotted with anti-GFP, anti-AKT or immunoprecipitated with anti-GFP followed by blotting with anti-GFP or anti-AKT. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 AKT activates TMEM175 in a catalytic activity-independent manner.
a, Schematic of the canonical AKT activation, with inhibitors used to target each step indicated, modified from previous publications23,80. A domain structure of AKT is in the bottom. AKT has a PH domain (PH), a kinase domain (KD) and a C-terminal tail regulatory domain (RD). b, Phosphorylation status of AKT and its two canonical targets GSK3β and PRAS40 following treatments similar to those used in the patch-clamp recording of TMEM175. Lysates from human TMEM175-transfected cells were probed with antibodies as indicated. Treatment conditions were: starvation (0 h, 4 h in HBSS) without (lanes 1 and 2) or with insulin (lane 3, 100 nM, 4 h) treatment afterwards, wortmannin (20 μM, 2 h) with and without SC79 (10 μM, 30 min) treatment afterwards (lanes 5 and 6), BKM120 (10 μM, 3 h; lane 8), GSK2334470 (20 μM, 3 h; lane 10), ARQ-092 (20 μM, 3 h; lane 12), MK2206 (20 μM, 3 h; lane 14), inhibitor VIII (10 μM, 3 h; lane 16), afuresertib (20 μM, 3 h; lane 18), AZD-5363 (20 μM, 3 h; lane 20), GSK690693 (10 μM, 4 h; lane 22). For gel source data, see Supplementary Fig. 1. c–i, Lysosomal IK was recorded from TMEM175-transfected HEK293T cells pretreated with wortmannin (c), BKM120 (d), GSK2334470 (e), GSK690693 (f), AZD-5363 (g) or afluresertib (h) as in b before lysosomal dissection for recording. Averaged IK sizes (at 100 mV) are shown in i. Data are mean ± s.e.m. Numbers of recordings are in parentheses. Our finding that SC79 fully activates TMEM175 in starved lysosomes was surprising but is also consistent with the idea that channel activation is independent of the kinase activity of AKT. Upon receptor activation by growth factors, PI3 kinases (PI3Ks) increase the concentration of PIP3 from PtdIns(4,5)P2 (PIP2)18. SC79 and the endogenous AKT activator PIP3 both bind to the PH domain of AKTs to induce large conformational changes that lead to the release of the PH domain from the catalytic domain and to an open state of the kinase19, which renders AKT accessible to phosphorylation at a key residue in the kinase domain (T308 in human AKT1) by PKD1 and another residue (S473) in the C-terminal regulatory domain by kinases such as mTORC2. Phosphorylation of T308 and S473 is required for the efficient activation of AKT kinase activity to phosphorylate downstream targets such as GSK3β and PRAS4018 (a). In nutrient-replete cells, TMEM175-associated AKT is at least partially phosphorylated at the two sites (Extended Data Fig. 4f). We used a panel of AKT inhibitors and mutants to probe the involvement of each of the steps in AKT function in TMEM175 activation. In starved cells and wortmannin-treated ones, AKT kinase activity is minimal because of the lack of phosphorylation of T308 on AKT by PKD1. Indeed, the activation of AKT kinase activity by SC79 on AKT isolated from starved cells requires the addition of PKD1 and ATP. Similarly, SC79 does not induce substantial AKT kinase activity (assayed with target phosphorylation) in wortmannin-treated cells19 (b). However, SC79 maximally activated TMEM175 in lysosomes isolated from starved cells (Fig. 2d, h) and in those from wortmannin-treated cells (c, i), without PKD1 or ATP in the bath, supporting that the SC79-induced channel activation does not require the kinase activity of AKT. Reduction of PIP3 by starvation and by inhibition of PI3K with wortmannin or BKM120 (a specific PI3K inhibitor)—treatments expected to keep AKT in a PH-in a closed state—both reduced the phosphorylation of AKT at T308 and S473 and that of the downstream AKT targets GSK3 (S9) and PRAS40 (T246) (b). The treatments also eliminated ITMEM175, which was restored by SC79 application (b–d, i). These data suggest that AKT activates channel after it switches from the PH-in closed state to the PH-out open state. To further test whether—after AKT switches to a PH-out open state—phosphorylation of T308 in AKT by PDK1 is required for ITMEM175, we inhibited PDK1 with GSK2334470. Although the inhibitor abolished the phosphorylation of AKT and its downstream targets, it had little effect on ITMEM175 (b, e, i). Catalytic and allosteric AKT inhibitors both block phosphorylation of downstream targets by AKT, but via different mechanisms18 (a). Unlike allosteric inhibitors, which probably keep AKT in a closed phosphorylation-resistant state and prevent the activation of TMEM175 (b, Fig. 2f), catalytic inhibitors such as those that compete with ATP binding in the activation loop are downstream of the phosphorylation of AKT T308 and S473 and inhibit AKT kinase activities in an open state. The ATP competitive inhibitors GSK690693, AZD-5363 and GSK2110183 (afuresertib) all inhibited AKT target phosphorylation (b), but—in contrast to the allosteric inhibitors (Fig. 2f)—had little effect on ITMEM175 (f–i). Together, the inhibitors targeting different steps of AKT activation suggest that conformational changes that lead to AKT in an open state, but not the downstream steps of the catalytic activities of AKT, are required for ITMEM175.
Extended Data Fig. 6 AKT determines the sensitivity of TMEM175 to starvation and activation by phosphatidylinositol lipids.
a–e, HEK293T cells (a, c–e) (lysosomal recording) or S2 cells (b) (whole-cell recording) were transfected with human TMEM175 with or without cotransfection of wild-type or mutant human AKT1. a, b, Experiments demonstrating that AKT with mutations in the kinase domain (K179M alone, T308A/S473A double mutant) are able to support ITMEM175 as well as wild-type AKT. Extended Data Figure 2 provides a comparison with those from S2 cells transfected with TMEM175 without human AKT1 cotransfection, in which very little currents were generated. A summary of current amplitudes (at −100 mV) is given in the bar graphs. The right panel in a shows the phosphorylation status of transfected AKT and AKT targets GSK3β and PRAS40. The human AKT1 used for transfection has an HA tag and a linker sequence, and can be distinguished from the endogenous AKT (indicated by green arrows) because of its slightly higher molecular weight. For gel source data, see Supplementary Fig. 1. In c–e, stv indicates cells starved in HBSS medium for 2 h before lysosomal dissection for recording. c, PIP3 (1 μM) was applied to the recording bath during some of the recordings (blue and red traces). d, e, ITMEM175 recorded from lysosomes with or without cotransfection of the E17K (d) or the D323A/D325A (DA) (e) AKT1 mutants, with or without starvation. PIP2 in d was applied at 1 μM. Data are mean ± s.e.m. Numbers of recordings are in parentheses. Arrows are used to indicate curves that overlap and are not easily distinguished. f, Positions of the AKT1 mutations used in this figure. We used AKT mutants to further test the nonessential role of kinase activity in ITMEM175. AKT1 with mutation at K179 (a key residue in the ATP- and Mg2+-binding pocket; K179M), or at T308 and S473 (residues key to AKT kinase activation; T308A/S473A) no longer phosphorylates its targets18,81. ITMEM175 from cells co-expressing the mutants, however, was similar to that co-expressing wild-type AKT (a). More importantly, when co-expressed with human TMEM175 in S2 cells, the kinase-inactive mutants fully supported ITMEM175 in a manner indistinguishable from that of the wild type (b). We then tested whether AKT conformational changes are sufficient to activate the channel. Under physiological conditions, PIP3 generated on the plasma membrane opens AKT by binding to the PH domain. In lysosomes from starved cells, PIP3 bath application activated ITMEM175 (c). The E17K mutation in the PH domain—commonly found in cancers—switches the lipid sensitivity of AKT from PIP3 to PIP2, a lipid that is abundant even during starvation82. Notably, ITMEM175 from cells cotransfected with AKT(E17K) was resistant to starvation, and, unlike that without cotransfection, was activated by PIP2 (d). Thus, AKT also determines the lipid sensitivity of TMEM175. The double mutation D323A/D325A of AKT1 disrupts the locking of the PH domain to the kinase domain and keeps AKT in a constitutively open state23,83. ITMEM175 from cells co-expressing this mutant was resistant to starvation, maximally activated and not further activated by SC79 (e).
Extended Data Fig. 7 Association of AKT to TMEM175 is required to maintain ITMEM175 after the channel is activated.
a, b, Development of peptide inhibitors that dissociate AKT from TMEM175 in the lysoKGF complex. a, Sequences of synthesized human TMEM175 peptides around the K336 region that interacts with AKT. Peptide 2 has a K336A mutation. b, To test the ability of each peptide to dissociate AKT from TMEM175, cell lysates from HEK293T cells cotransfected with GFP-tagged human TMEM175 and HA-tagged human AKT1 were incubated with each peptide (10 μM) for 2 h before immunoprecipitation with anti-GFP to bring down TMEM175 and immunoblotting with anti-HA to detect the associated AKT1. Immunoprecipitation without peptide incubation (lane 1) was used as a control. Whole-cell lysate was also blotted with anti-HA and anti-GFP for input control. For gel source data, see Supplementary Fig. 1. The results show that when applied in cell lysates containing the TMEM175–AKT complex, peptides as short as 10 amino acids (peptide 5) in the K336 region were sufficient to dissociate AKT from the preassembled AKT–TMEM175 complex in the immunoprecipitation assay. As a control, K336A mutation (peptide 2) abolished the dissociation ability of the peptide. c, d, Lysosomal IK were recorded from TMEM175-transfected HEK293T cells (c) and mouse hippocampal neurons (d). After SC79 application to maximally activate lysoKGF, peptide inhibitor (10 μM) was added to the bath and currents were recorded for about 10 min until they reached steady states. The K336A mutant peptide 2 was used as a negative control. Averaged IK sizes (at 100 mV) are shown in the bar graphs. In c subpanel ‘peptide 2’, arrows are used to indicate curves that overlap and are not easily distinguished. Blue, from bath containing K+ and SC79 before peptide application; red, from bath containing K+ and SC79 after peptide application. Data are mean ± s.e.m. Numbers of recordings are in parentheses. *P ≤ 0.05. P values (unpaired two-tailed t-tests) (K+SC79 vs K+SC79 + peptide groups) are as follows. In c, P = 0.8447 for peptide 2, P < 0.0001 for the other treatments; in d, P = 0.0011 for peptide 5. The results show that after the channel was maximally activated by SC79, bath application of AKT-association peptide inhibitors—but not the one with the K336A point mutation—diminished both heterologously expressed ITMEM175 in HEK293T cells and the native currents in neurons. Given that the bath contained no phosphatase, the action of the peptides is unlikely through a dephosphorylation of the channel target but is rather through the dissociation of AKT from TMEM175—further supporting the idea that it is the conformational change of AKT, and not target phosphorylation, that activates the channel. e, A model for TMEM175 activation by AKT conformational changes. TMEM175 without AKT or associated with inactive AKT in its PH-in closed conformation is closed. The channel opens upon AKT conformational changes triggered by physiological activators such as extracellular growth factors, lipids, or synthetic small molecules such as SC79. The gating mechanism does not require AKT kinase activity. AKT mutations found in cancers that alter the interaction between the PH and kinase domains also alter TMEM175 channel property.
Extended Data Fig. 8 Knock-in mice to model common human variants p.M393T and p.Q65P.
a–d, Experiments demonstrating that the TMEM175(I390T) substitution in mice—corresponding to M393T in human TMEM175—leads to reduced lysosomal IK, accelerated spreading of pathogenic α-syn and increased neuronal damage. a, Sanger sequencing of genomic DNA PCR products from wild-type, heterozygous and homozygous TMEM175(I390T) knock-in mice in the I390-coding region. Residue I390 in mouse TMEM175 is equivalent to M393 in human TMEM175, as shown in the sequence alignment on the right. b, IK recorded from wild-type (+/+, wild type), heterozygous (I390T/+, het) and homozygous (I390T/I390T, hom) knock-in mouse midbrain neurons. Bar graphs show averaged IK (at 100 mV). Data are mean ± s.e.m. Numbers of recordings are in parentheses. c, Neuronal damages in wild-type, het and hom midbrain neurons treated with MPP+ with concentrations as indicated added to culture medium for 12 h and those followed by incubation with insulin (100 nM in DMEM) for 3 h. Damage index was calculated as the ratio of damaged area to the total neuronal area. d, Cultured wild-type and TMEM175(I390T) (hom) midbrain neurons were seeded with mouse α-syn PFFs and immunostained with anti-pSer129 α-syn (pSyn, red) two weeks later. β-Tubulin (green) was costained to highlight neuronal processes. DAPI (blue) nuclear staining was used to identify number of cells. Left two subpanels, representative images. Right, pSyn signals normalized to number of cells (n = 8 coverslips each). For c, d, see Fig. 5, Extended Data Fig. 10 for more details. P values in c are as follows (unpaired two-tailed t-tests, compared to wild type): 0 µM MPP+, P = 0.9318 (het), P = 0.2516 (hom); P < 0.0001 for all the other comparisons. The P value in d (t-test, wild type versus hom) is P < 0.0001. e–h, Characterization of the rs34884217 (p.Q65P) variant. e, Genomic structures of the human and mouse TMEM175 genes around the rs34884217 SNP region. The A-to-C variation in human and the corresponding substitution in the knock-in mouse line are indicated by arrows. f, Agarose gel electrophoresis showing RT–PCR products of the whole open reading frame cDNA (about 1.5 kb) amplified from wild-type and knock-in mouse brains. For gel source data, see Supplementary Fig. 1. g, Sanger sequencing results of the RT–PCR products in f, demonstrating that the A-to-C substitution in mice leads to a Q65P coding change. h, Sequence alignments around Q65 in the I-TM1–TM2 linker. Q65P is located at the luminal side of the channel entrance.
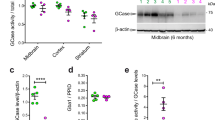
Extended Data Fig. 9 TMEM175 contributes to lysosomal pH stability and membrane integrity, regulates lysosomal enzyme activity and controls organelle fusion during autophagy.
a, Lysosomal pH from wild-type and TMEM175-knockout cultured mouse hippocampal neurons with or without starvation (overnight in DMEM). Distribution of pH values are in the first two subpanels. Average values are in the bar graph. Numbers of lysosomes analysed are in parentheses. b, Comparison of mature CatD levels using anti-CatD antibody. Lysate from cultured wild-type or TMEM175-knockout hippocampal neurons, with or without starvation, were probed with anti- CatD, or anti-β-actin for loading control. The mature form of CatD (bottom band) is cleaved from pro-CatD (top band) and the cleavage is pH-dependent. Left, representative blot. Right, signal density of mature CatD normalized to that of pro-CatD (bottom) or to that of β-actin (top). n = 5 repeats. *P ≤ 0.05. P values (unpaired two-tailed t-test, wild type vs knockout) are as follows: normalized to β-actin, P = 0.0119 (without starvation), P = 0.0011 (with starvation); normalized to pro form, P = 0.0002 (without starvation), P = 0.0015 (with starvation). For gel source data, see Supplementary Fig. 1. c, Comparison of mature CatD levels using BODIPY FL pepstatin A (a fluorescent cathepsin indicator that binds to CatD in a pH-dependent manner). Cultured wild-type and TMEM175-knockout hippocampal neurons, with or without starvation, were treated with BODIPY FL pepstatin A (1 μM, 1 h) and imaged for pepstatin A and nuclei. Left, representative images of pepstain A (green), DAPI (blue) and merge between the two. Right, pepstain A signals normalized to numbers of cells (n, number of coverslips imaged). d, Autophagosome–lysosome fusion assay with an mCherry- and GFP-tagged LC3 (tandem tag, illustrated) transfected into wild-type and TMEM175-knockout hippocampus neurons with or without starvation. Left, representative images for GFP (indicative of autophagosome puncta before fusion with lysosome; GFP is invisible after fusion with lysosome because of its pH sensitivity), RFP (indicative of total number of autophagosome puncta before and after phagosome–lysosome fusion; RFP is visible under both neutral and acidic pH) and DAPI (nucleus), and merge among the three. Bar graph indicated averaged numbers of GFP and RFP puncta upon treatments as indicated (middle) and numbers of GFP-positive puncta normalized to those of RFP-positive ones (right). Number of cells analysed are in parentheses. e, Experiments demonstrating that deficiency in lysoKGF leads to accelerated spreading of pathogenic α-syn and lysosomal permeability in neurons. Cultured wild-type and TMEM175-knockout hippocampal neurons were seeded with mouse α-syn PFFs and costained with anti-pSer129 α-syn (pSyn, red) and anti-galectin 3 (green) two weeks later. DAPI (blue) nuclear staining was used to identify the number of cells. Representative images are in the left panels and averaged pSyn and galectin 3 signal density (normalized to number of cells) are in the right bar graph (n = 6 coverslips each). ***P ≤ 0.001. P values (unpaired two-tailed t-tests; knock out versus wild type) are P < 0.0001 for α-syn and galectin 3. In the bar graphs, data are represented as mean ± s.e.m.
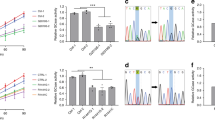
Extended Data Fig. 10 LysoKGF protects mouse neurons from stress-induced damage in a gene-dosage-dependent manner, and a loss-of-function variant in the human TMEM175 gene is nominally associated with increased rates of cognitive and motor function decline in patients with Parkinson’s disease.
a–e, Cultured mouse neurons were challenged with MPP+ toxin, B27 deprivation (in DMEM) and/or H2O2 treatment. Images were taken for damage analysis. Blebs and fragmentation in neuronal processes were counted as damage, as indicated by arrows in representative pictures in a. The damage index was calculated as the ratio of damaged area to the total neuronal area. a, Treatment of wild-type and TMEM175-knockout midbrain neurons with MPP+ added to culture medium for 12 h, with concentrations as indicated. Left, representative images of neurons with bleb damages indicated by arrows. Right, damage index. b, c, Hippocampal (b) or midbrain (c) neurons were starved in DMEM for duration as indicated. H2O2 (100 μM) treatment for the period indicated was carried out in DMEM after starvation. d, Effects of insulin. In the group of neurons treated with 14-h starvation and insulin, without H2O2, neurons were starved for 14 h in DMEM followed by incubation with insulin (100 nM in DMEM) for 3 h. In the groups treated with H2O2, neurons were starved in DMEM for 0 or 14 h, followed by a 3-h incubation with or without insulin (100 nM). During the last hour of incubation, H2O2 (100 μM) was added to the medium. e, Comparison between wild-type and TMEM175(Q65P)-carrying neurons with treatments similar to those in c. In the bar graphs, data are mean ± s.e.m.; numbers of neurons analysed are in parentheses. *P ≤ 0.05. P values (unpaired two-tailed t-tests) are as follows. In a, wild type versus knockout: P < 0.0001 for all comparisons. In b, wild type versus knockout: P = 0.0192 (0 h, starved), P = 0.0004 (3 h, starved), P < 0.0001 for the other comparisons. In c, wild type versus knockout: P = 0.0017 (0 h + H2O2), P < 0.0001 for the other comparisons. In d, compared with wild type: P = 0.264 (HET, 0 h), P < 0.0001 for the other comparisons. In e, compared with wild type: P = 0.8121 (+/Q65P, 0 h), P = 0.545 (Q65P/Q65P, 0 h), P = 0.0935 (+/Q65P, 3 h), P = 0.2012 (Q65P/Q65P, 3 h), P < 0.0001 for the other comparisons. f, Clinical effect of the rs34311866 (p.M393T) variant in the human TMEM175 on longitudinal course of Parkinson’s disease. Longitudinal effects for carriers of no (blue), one (green), or two (red) p.M393T variants are shown for mixed-effects linear regression models without multiple testing correction; 95% confidence intervals are indicated by coloured bands. In the UPenn cohort, p.M393T carriers had faster cognitive decline (two-tailed P = 0.005, as captured by lower DRS-2 scores) and faster motor decline (two-tailed P = 0.032, higher UPDRS-3 scores). In the PPMI cohort, p.M393T carriers trended (two-tailed P = 0.066) towards faster motor decline, although no statistical significance was reached (presumably because of shorter observation period, of 5 years after diagnosis). As TMEM175 variants are associated with neurodegenerative diseases and AKT is known to protect neurons against stress-induced damage25,35,36, we tested whether lysoKGF protects neurons. To do this, we used a neuronal culture model in which damage induced by insults such as the neurotoxin MPTP, H2O2 and nutrient removal can be quantified well37,38,39. MPTP interferes with mitochondrial metabolism, inducing parkinsonism in humans and in animal models84. Consistent with previous findings85,86, incubating midbrain neurons with MPP+ (the active form of the neurotoxin) led to perturbations and beading of neuronal processes in a dose-dependent manner. When the size of such damaged areas normalized to the total neuronal area (that is, damage index) was quantified, TMEM175-knockout neurons exhibited much more severe damage compared to wild type (a). Both nutrient removal and reactive oxygen species (such as H2O2) cause neuronal damage39. Under each combination of H2O2 treatment after B27 removal, TMEM175-knockout neurons exhibited more severe damage than wild type (b, c). Growth factors such as insulin are known to mitigate neuronal damage but the mechanisms are not well-understood87. In wild-type neurons, insulin reduced the damage induced by B27 removal and/or H2O2 treatment. Notably, this protection was nearly absent in TMEM175-knockout neurons (d), which suggests an essential role for TMEM175. In heterozygous neurons, the degree of damage was intermediate between that of wild-type and TMEM175-knockout neurons (d). Thus, Tmem175 gene dosage has an incremental effect on neuronal protection. Decreasing ITMEM175 by about 50% is sufficient to compromise the protection. In support of this finding, neurons from TMEM175(I390T) knock-in mice were also more prone to MPP+-induced damage than were neurons from wild-type mice (Extended Data Fig. 8c). We also tested whether p.Q65P provides additional protection. When challenged with B27 deprivation and H2O2, neurons from both heterozygous (Q65P/+) and homozygous (Q65P/Q65P) mice had much less damage than those from wild type (e). The difference was even more notable between TMEM175(Q65P)-carrying neurons and TMEM175-knockout neurons. For example, after a 14-h starvation the TMEM175-knockout neurons had 27% damage and wild-type neurons had 15%—but those from Tmem175Q65P/+ mice had only 6%. Similarly, H2O2-induced damages were mitigated in TMEM175(Q65P)-carrying neurons (c, e). These data suggest that p.Q65P is neuroprotective and constitutes a potential mechanism that underlies a reduced susceptibility to developing Parkinson’s disease.
Supplementary information
Supplementary Figure
This file contains original gel pictures used to make the figure.
Source data
Rights and permissions
About this article
Cite this article
Wie, J., Liu, Z., Song, H. et al. A growth-factor-activated lysosomal K+ channel regulates Parkinson’s pathology. Nature 591, 431–437 (2021). https://doi.org/10.1038/s41586-021-03185-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-03185-z
This article is cited by
-
Drosophila TMEM63 and mouse TMEM63A are lysosomal mechanosensory ion channels
Nature Cell Biology (2024)
-
Transmembrane proteins with unknown function (TMEMs) as ion channels: electrophysiological properties, structure, and pathophysiological roles
Experimental & Molecular Medicine (2024)
-
Clinical biomarkers for Lewy body diseases
Cell & Bioscience (2023)
-
Electrophysiological damage to neuronal membrane alters ephaptic entrainment
Scientific Reports (2023)
-
Metal-organic framework template-guided electrochemical lithography on substrates for SERS sensing applications
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.