Abstract
The formation of arteries is thought to occur by the induction of a highly conserved arterial genetic programme in a subset of vessels that will later experience an increase in oxygenated blood flow1,2. The initial steps of arterial specification require both the VEGF and Notch signalling pathways3,4,5. Here, we combine inducible genetic mosaics and transcriptomics to modulate and define the function of these signalling pathways in cell proliferation, arteriovenous differentiation and mobilization. We show that endothelial cells with high levels of VEGF or Notch signalling are intrinsically biased to mobilize and form arteries; however, they are not genetically pre-determined, and can also form veins. Mechanistically, we found that increased levels of VEGF and Notch signalling in pre-arterial capillaries suppresses MYC-dependent metabolic and cell-cycle activities, and promotes the incorporation of endothelial cells into arteries. Mosaic lineage-tracing studies showed that endothelial cells that lack the Notch–RBPJ transcriptional activator complex rarely form arteries; however, these cells regained the ability to form arteries when the function of MYC was suppressed. Thus, the development of arteries does not require the direct induction of a Notch-dependent arterial differentiation programme, but instead depends on the timely suppression of endothelial cell-cycle progression and metabolism, a process that precedes arterial mobilization and complete differentiation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA-seq data can be viewed at the Gene Expression Omnibus (GEO) under accession number GSE158731. We used in this study the MsigDB as indicated above. All other data supporting the study findings are available from the corresponding author upon request. This includes additional raw data such as unprocessed original pictures and independent replicates, which are not displayed in the Article but are included in the data analysis in the form of graphs. Source data are provided with this paper.
References
Red-Horse, K. & Siekmann, A. F. Veins and arteries build hierarchical branching patterns differently: bottom-up versus top-down. BioEssays 41, e1800198 (2019).
Simons, M. & Eichmann, A. Molecular controls of arterial morphogenesis. Circ. Res. 116, 1712–1724 (2015).
Phng, L. K. & Gerhardt, H. Angiogenesis: a team effort coordinated by notch. Dev. Cell 16, 196–208 (2009).
Benedito, R. & Hellström, M. Notch as a hub for signaling in angiogenesis. Exp. Cell Res. 319, 1281–1288 (2013).
Lawson, N. D., Vogel, A. M. & Weinstein, B. M. Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell 3, 127–136 (2002).
Pontes-Quero, S. et al. High mitogenic stimulation arrests angiogenesis. Nat. Commun. 10, 2016 (2019).
Benedito, R. & Duarte, A. Expression of Dll4 during mouse embryogenesis suggests multiple developmental roles. Gene Expr. Patterns 5, 750–755 (2005).
Duarte, A. et al. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 18, 2474–2478 (2004).
Gale, N. W. et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl Acad. Sci. USA 101, 15949–15954 (2004).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 (2006).
Su, T. et al. Single-cell analysis of early progenitor cells that build coronary arteries. Nature 559, 356–362 (2018).
Fang, J. S. et al. Shear-induced Notch-Cx37-p27 axis arrests endothelial cell cycle to enable arterial specification. Nat. Commun. 8, 2149 (2017).
Pontes-Quero, S. et al. Dual ifgMosaic: a versatile method for multispectral and combinatorial mosaic gene-function analysis. Cell 170, 800–814 (2017).
Benedito, R. et al. The Notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell 137, 1124–1135 (2009).
Wu, B. et al. Endocardial cells form the coronary arteries by angiogenesis through myocardial-endocardial VEGF signaling. Cell 151, 1083–1096 (2012).
Chang, A. H. et al. DACH1 stimulates shear stress-guided endothelial cell migration and coronary artery growth through the CXCL12-CXCR4 signaling axis. Genes Dev. 31, 1308–1324 (2017).
Chen, H. I. et al. The sinus venosus contributes to coronary vasculature through VEGFC-stimulated angiogenesis. Development 141, 4500–4512 (2014).
Aranguren, X. L. et al. Unraveling a novel transcription factor code determining the human arterial-specific endothelial cell signature. Blood 122, 3982–3992 (2013).
Ehling, M., Adams, S., Benedito, R. & Adams, R. H. Notch controls retinal blood vessel maturation and quiescence. Development 140, 3051–3061 (2013).
Lee, T., Yao, G., Nevins, J. & You, L. Sensing and integration of Erk and PI3K signals by Myc. PLOS Comput. Biol. 4, e1000013 (2008).
Hasan, S. S. et al. Endothelial Notch signalling limits angiogenesis via control of artery formation. Nat. Cell Biol. 19, 928–940 (2017).
Pitulescu, M. E. et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol. 19, 915–927 (2017).
Sissaoui, S. et al. Genomic characterization of endothelial enhancers reveals a multifunctional role for NR2F2 in regulation of arteriovenous gene expression. Circ. Res. 126, 875–888 (2020).
Gaudino, M. et al. Radial-artery or saphenous-vein grafts in coronary-artery bypass surgery. N. Engl. J. Med. 378, 2069–2077 (2018).
Fujita, M. & Sasayama, S. Coronary collateral growth and its therapeutic application to coronary artery disease. Circ. J. 74, 1283–1289 (2010).
Kisanuki, Y. Y. et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242 (2001).
Wang, Y. et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature 465, 483–486 (2010).
Claxton, S. et al. Efficient, inducible Cre-recombinase activation in vascular endothelium. Genesis 46, 74–80 (2008).
Koch, U. et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J. Exp. Med. 205, 2515–2523 (2008).
Han, H. et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int. Immunol. 14, 637–645 (2002).
de Alboran, I. M. et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55 (2001).
Huang, C. Y., Bredemeyer, A. L., Walker, L. M., Bassing, C. H. & Sleckman, B. P. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur. J. Immunol. 38, 342–349 (2008).
Fernández-Chacón, M. et al. iSuRe-Cre is a genetic tool to reliably induce and report Cre-dependent genetic modifications. Nat. Commun. 10, 2262 (2019).
Lim, R. et al. Deubiquitinase USP10 regulates Notch signaling in the endothelium. Science 364, 188–193 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10 (2011).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Liberzon, A. et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011).
Scheurer, S. B., Rybak, J. N., Rösli, C., Neri, D. & Elia, G. Modulation of gene expression by hypoxia in human umbilical cord vein endothelial cells: A transcriptomic and proteomic study. Proteomics 4, 1737–1760 (2004).
Weigand, J. E., Boeckel, J. N., Gellert, P. & Dimmeler, S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One 7, e42697 (2012).
Chu, V. T. et al. Efficient generation of Rosa26 knock-in mice using CRISPR/Cas9 in C57BL/6 zygotes. BMC Biotechnol. 16, 4 (2016).
Acknowledgements
Research in the Benedito laboratory was supported by the European Research Council (ERC) Starting Grant AngioGenesHD (638028), the CNIC Intramural Grant Program Severo Ochoa (11-2016-IGP-SEV-2015-0505), and the Ministerio de Ciencia y Innovación (MCIN SAF2013-44329-P, RYC-2013-13209 and SAF2017-89299-P). The CNIC is currently supported by MCIN and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (SEV-2015-0505). Research in the Potente laboratory was supported by the Max Planck Society, the ERC Consolidator Grant EMERGE (773047), the Deutsche Forschungsgemeinschaft (SFB 834), and the Foundation Leducq Transatlantic Network. W.L. received a Marie Curie FP7 COFUND CNIC fellowship. M.F.-C. and I.G.-G. were supported by PhD fellowships from Fundación La Caixa (CX_E-2015-01 and CX-SO-16-1, respectively) and S. M. by the Austrian Science Fund (FWF) project J4358. We thank S. Bartlett and S. Rocha for English editing; J. L. de La Pompa and D. Macgrogan for scientific input and the CNIC Transgenesis, Microscopy, Genomics and Bioinformatic units. We also thank M. Yanagisawa, F. Radtke, R. H. Adams, M. Fruttiger, F. Alt, B. Sleckman and T. Honjo for sharing the Tie2-cre, Dll4floxed, Cdh5(PAC)-creERT2, Pdgfb-icreERT2-ires-egfp, Mycfloxed, GFP-Myc and Rbpjfloxed mice, respectively.
Author information
Authors and Affiliations
Contributions
W.L. and R.B. designed most of the experiments, interpreted results and wrote the manuscript. I.G.-G. engineered the DNA constructs, validated the iFlpMTomato-cre/MYfp and Apln-FlpO mice, and gave input on genetic mosaics analysis. M.F.-C. supported RNA-seq, FACS, qRT–PCR and iSuRe-cre analysis. L.G.-O. performed mouse aortas dissection, immunostaining and analysis. V.C.-G. and M.S.S.-M. gave general technical assistance, managed the mouse colonies and breedings, and established some of the ES cell lines used to generate mouse models. S.M., J.A. and M.P. designed and performed experiments with endothelial cell lines, interpreted results and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Andreas Fischer, Holger Gerhardt and Kristy Red-Horse for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
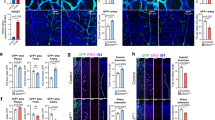
Extended Data Fig. 1 Multispectral genetic mosaics to analyse the proliferation and arteriovenous fate of heart and retina ECs.
a, Schematic of Dual ifgMosaic technology to multispectrally barcode cells with different combinations of chromatin (iChr2-Mosaic) or membrane (iMb2-Mosaic) tagged fluorescent proteins. b, Combined or individual channels of multispectral confocal imaging of the heart-surface coronary vasculature and sections. c, iChr-Notch-Mosaic allele targeted to the Rosa26 locus. In cells expressing Cre, the allele undergoes stochastic recombination, generating a mosaic of cells expressing either H2B-Cherry (control wild-type cells with Cherry FP bound to chromatin/nuclei), H2B-GFP-2A-DN-MAML1 (GFP labels the nuclei of cells with induced downregulation of Notch/Rbpj signalling) or HA-H2B-Cerulean-2A-N1ICDP (Cerulean containing the HA epitope labels the nuclei of cells with induced upregulation of NOTCH1 signalling). d, qRT–PCR analysis of Notch target-genes in H2B-GFP+ and H2B-Cherry+ ECs obtained from iChr-NotchTie2-Mosaic E15.5 hearts by FACS. e, Survival rate of adult iChr-NotchTie2-Mosaic mice follows expected Mendelian frequencies indicating no lethality. f, g, Confocal images of iChr-NotchNfatc1-Mosaic embryonic hearts sections showing the localization and relative frequency of ECs with distinct Notch signalling levels. h, Quantification of experiments in f and g showing how the relative percentage of each type of cell changes throughout development according to their location. i, Proliferation (EdU+) of coronary ECs (ICAM2+/ERG+ in myocardium) and endocardium (EMCN+/ERG+) at E12.5 and E15.5. j, k, Analysis of cell proliferation in iChr-NotchTie2-Mosaic ECs. l, Illustration showing the iFlpMTomato-cre/MYfp mosaic allele and expected outcomes in cells expressing Flp/FlpO recombinase. m, qRT–PCR analysis of Rbpj mRNA levels in CD31+/MbYFP+ and CD31+/MbTomato+ ECs collected by FACS from Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjf/f E15.5 hearts. Residual Rbpj expression is due to the FACS and qRT–PCR assay as previously described33 (see Methods). n, Confocal images and quantification of postnatal day 6 (P6) retinas showing the localization and frequency of MTomato+ (RbpjKO) and MYFP+ (wild-type) cells in retina arteries (A), veins (V) and intermediate capillaries. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 2 Ectopic Notch activation is compatible with vein development and impairs coronary artery development.
a, Genetic construct used to generate mice with Cre-dependent conditional expression of MbTomato, H2B-GFP and N1ICDP in Tie2-Cre+ coronary vessels which results in increased expression of canonical Notch target genes. b, Bright-field images of E12.5 and E15.5 embryos and hearts showing the appearance of microcirculatory defects at E15.5. c, Statistical analysis of mouse survival rate per stage indicates that most mutant mice do not pass embryonic stage E16.5. d, Cardiovascular development defects in N1ICDPTie2-cre hearts may result in tissue hypoxia and the observed upregulation of Vegf mRNA. e, f, Myocardial EC proliferation analysis. g, h, CD31 staining of control and N1ICDPTie2-cre heart sections showing vascular defects and thinner myocardial wall (inset) in mutant hearts. i–m, Analysis of the heart surface coronary vein diameter and plexus development and density. n, Confocal images showing immunostaining of the heart surface venous plexus for the arterial marker CX40. ECs with high Notch activity (Tomato+) do not express CX40 and are able to form veins. o, Confocal images of E14.5 heart veins showing that Tomato/N1ICDP+ cells (yellow arrowheads) express EPHB4 like wild-type cells (white arrowheads). p, Confocal images of E14.5 heart veins showing that GFP+/N1ICDP+ cells (yellow arrowheads) express COUP-TFII like GFP−/wild-type cells (white arrowheads). q, Chart showing the quantification of COUP-TFII immunosignals intensity in 280 GFP+ and 280 GFP− cells present in the veins of 3 independent N1ICDPTie2-cre hearts (2 veins per heart were quantified). r, Schematic of the mouse postnatal retina vascular development, in which tamoxifen was induced at P1, when the vessels start to grow, and analysis was carried at P6. s–v, Cells with induced high Notch activity (MbTomato+ or H2B-GFP+, yellow arrowheads) can differentiate and form veins like wild-type cells (white arrowheads). The Tomato+/N1ICDP+ cells found in veins do not express the arterial marker CX40, and instead express the venous markers VEGFR3, COUP-TFII and EPHB4. w, Quantification of signals (in pictures like shown in s–v) indicate no change in the expression of arteriovenous genes. x, Analysis of coronary artery development. y, z, Confocal images of heart sections and quantifications of immunosignals in the myocardium showing an increase in CX40, DLL4 and SM22α in N1ICDPTie2-cre myocardium vessels. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 3 Overactivation of Notch in PDGFB+ coronary ECs, but not in NFATC1+ endocardium, compromises coronary vessel and heart development.
a, Schematic of the inducible Notch gain-of-function allele, that once crossed with the Pdgfb-icreERT2-ires-egfp allele enables the co-expression of Mbtomato, H2B-GFP and N1ICDP in PDGFB+ coronary ECs. b, Confocal picture showing the strong expression of the Pdgfb-icreERT2-ires-egfp allele in coronary vessels of the myocardium (m), but not in the EMCN+ endocardium (e). c, Stereomicroscope analysis of mutant embryos and control littermates showing growth retardation at E16.5. d, Immunostaining for the reporter MbTomato shows the induction of the allele in Pdgfb-icreERT2-ires-egfp+ myocardium vessels (ICAM2+) but not endocardium. e, g, Thinner myocardial wall and vascular defects in N1ICDPPdgfb-creERT hearts. f, h, Malformation and reduced coronary arterial diameter in N1ICDPPdgfb-creERT hearts. i–k, Reduced coronary vessel density and vein diameter in N1ICDPPdgfb-creERT hearts. l, m, Expression of the arterial proteins DLL4 and CX40 in control and N1ICDPPdgfb-creERT coronary vessels. n, o, Reduced proliferation (ERG+/EdU+) of N1ICDPPdgfb-creERT subepicardium venous vessels (whole-mount and sections). p, Induction of N1ICDP expression (MbTomato+) in the NFATC1+ endocardium and its lineages does not cause major cardiovascular development defects at E15.5. q–t, Comparative analysis of coronary artery and vein development in control and N1ICDPNfatc1-cre hearts. Note the absence of GFP+/N1ICDP+ cells in veins derived from Nfatc1-cre lineage in N1ICDPNfatc1-cre mice. u, Haematoxylin and eosin staining of 7-month-old hearts of control and N1ICDPNfatc1-cre mice. Mutant hearts are larger. Quantifications of heart weight per body weight and left ventricle (LV) fraction shortening are shown. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 4 Induction of cell-cycle and metabolic pathways in coronary ECs 24 h after Dll4 deletion.
a, List of the 50 most up- and downregulated genes (from a total of 3,360) after Dll4 deletion in coronary ECs for 24 h. b, c, List of dysregulated genes within a selected enriched gene set (gene set enrichment analysis, GSEA). d, Fold changes for a selected list of genes previously shown to be upregulated when HUVECs are exposed to hypoxia40,41. e, Vegf mRNA levels do not change in hearts from Dll4KO-Pdgfb-24h (E14.5) and RbpjKO-iEC (E15.5) embryos, suggesting the absence of cardiac hypoxia, despite the existence of endothelial hypoxia. f, The group of genes belonging to the sprouting angiogenesis GSEA pathway are not differentially regulated. g, The expression of most genes previously found to be upregulated in tip cells is not increased after the loss of DLL4–NOTCH signalling. h, Only 7 genes are significantly dysregulated in Dll4Het-Pdgfb-24h samples. Data shown as mean ± s.d. *P < 0.05, Benjamini and Hochberg adjusted.
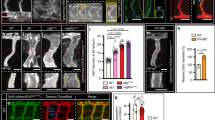
Extended Data Fig. 5 DLL4 signalling dose is essential for coronary proliferation and arterialization but not sprouting or connection to the aorta.
a, Dll4 deletion in Pdgfb-icreERT2-ires-egfp+ coronary ECs for 24 h (from E11.5–E12.5 or E13.5–E14.5) does not induce endothelial sprouting, but slightly reduces the mean density of heart subepicardial vessels at E14.5. b–e, The coronary plexus forming the coronary artery connects (yellow arrows) to the main aorta of control, Dll4Het-iEC and Dll4KO-iEC hearts. However, despite no defects in the connection, the calibre of the coronary artery stem is reduced and most Dll4Het-iEC hearts and all Dll4KO-iEC hearts do not develop proper coronary arteries with CX40+ signals (pink arrows) at E14.5. f, At E17.5, the surviving Dll4Het-iEC embryos also show a major developmental defect or delay in coronary artery development. Of note, the DLL4 heterozygous haploinsufficiency is variable and more pronounced on the C57BL/6J background used in this study, as previously reported8,9. g, h, Dll4 and Rbpj deletion reduces the frequency of EdU+ ECs in subepicardium vessels. i, Dll4 deletion increases p-ERK activity in subepicardium vessels. j, k, Dll4 and Rbpj deletion increases the frequency of p21+/ERG+ ECs in subepicardium vessels. l, m, Dll4 and Rbpj deletion increases the frequency of proliferation (EdU+/ERG+) in arterial zone coronary vessels. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 6 Regulation of MYC by DLL4–NOTCH signalling in vivo and in vitro.
a, MYC–GFP fusion protein expression is upregulated in coronary vessels (ICAM2+) after inducing heterozygous Dll4 deletion. b, Comparison of MYC protein expression in wild-type and Rbpj mutant ERG+ ECs. c, Western blot analysis showing no major changes in MYC protein levels after stimulation of HUVECs with DLL4 ligands for 6 or 24 h, in the presence or absence of the Notch inhibitor DBZ. d, qRT–PCR analysis showing significant changes of Notch target genes (Dll4, Hey1, Hey2, Hes1), but not Myc and Odc1, after stimulation of HUVECs with DLL4 ligands for 8 h in the presence or absence of the Notch inhibitor DBZ. e–h, Western blot and qRT–PCR analysis showing that an even stronger activation of Notch signalling (for 24 h, e and f; and 48 h g and h) and its downstream target genes (HEY1, HEY2, NRARP and HES1) in HUVECs overexpressing Dox-inducible N1ICD-V5 has relatively minor or non-significant effects on MYC expression. ODC1 expression is 40% decreased. i, Whole-mount analysis of control and MYC mutant hearts showing that global MYC deletion compromises the development of coronary vessels and consequently arteries (CX40+). j, Sectional analysis of control and MYC mutant hearts showing a severe depletion of coronary vessels in the myocardium which causes a reduction in its thickness. k, Mosaic induction of Mycflox/flox Cdh5-creERT2 iSuRe-cre mice shows that MYC-null ECs (MbTomato-2A-Cre+) form well coronary arteries, in contrast to hearts with full induction of Myc deletion. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1. For western blot gel source data, see Supplementary Fig. 1. Western blot controls (tubulin) were run on separate gels, owing to overlapping size with MYC, as sample processing controls.
Extended Data Fig. 7 Loss of Myc rescues the proliferation and arterialization defects caused by Rbpj or Dll4 deletion.
a, Sectional analysis of control and Rbpj/MycDKO-iEC iSuRe-cre mutant hearts showing that MbTomato-2A-Cre-expressing cells (from the induced iSuRe-cre allele) form CX40+ coronary arteries. b, qRT–PCR analysis showing the efficient deletion of Rbpj and Myc in Rbpj/MycDKO-iEC mutant cells. Note residual expression detected is due to the FACS and qRT–PCR assay as previously described33 (see Methods). c, Confocal pictures of the heart deeper arterial zone and quantifications showing that the additional loss of Myc rescues the proliferation (ERG+/EdU+) defects of RbpjKO ECs, rescuing also arterial development. d–f, Confocal pictures and quantifications showing that loss of Myc rescues the proliferation and arterial development defects induced by hemizygous loss of Dll4. g, h, The additional use of the iSuRe-cre allele, ensures that Tomato-2A-Cre expressing cells have deletion of all floxed alleles (in this case Dll4 and Myc). These cells do not form arteries on a Dll4floxed background but yes on a Dll4/Mycfloxed background. Note that Tomato- cells also express Cdh5-creERT2, which also efficiently deletes Dll4 and Myc, independently of recombination of the iSuRe-cre transgene. i, Arterial development in control, RbpjKO-iEC and Rbpj/MycDKO-iEC P6 retinas. Tamoxifen was injected at P2 and P3. Data shown as mean ± s.d. ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 8 Gene expression signatures of Dll4/Myc and Rbpj/Myc mutant coronary vessels.
a–l, qRT–PCR analysis of the indicated genes in the samples mentioned in a. Each dot in a chart column bar represents the relative measure in a sample of cells obtained by FACS from an entire litter containing several Pdgfb-icreERT2-ires-egfp+ hearts. Charts in e, h, j and l represent the fold change of all genes belonging to the indicated group and shows that the additional deletion of Myc rescues the expression of all sets of genes to control sample levels. m–o, qRT–PCR analysis of the indicated genes in the samples mentioned in m. Each dot in a chart column bar represents the relative measure in a sample of cells obtained by FACS from an entire litter containing several Tomato− and Tomato+ hearts. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. For statistics, see Supplementary Data 1.
Extended Data Fig. 9 iFlpMTomato-cre/MYfp genetic mosaics reveal the long term fate of RbpjKO and Rbpj/MycDKO ECs in different organs.
a, Schematic of the Apln-FlpO and iFlpMTomato-cre/MYfp alleles used to induce full loss-of-function genetic mosaics in the mouse heart, retina, liver and aorta. b, Lineage qRT–PCR analysis of Tomato+ and YFP+ cells isolated from Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjf/f and Apln-FlpO; iFlpMTomato-cre/MYfp;Rbpjf/f;Mycf/f E15.5 hearts. Note that Apln-FlpO recombines the alleles at an early stage and that mutant cells differentially segregate throughout development, leading to gene expression changes at E15.5 that may reflect not only the regulation by Rbpj and Myc (shown in Extended Data Fig. 8) but also their segregational arteriovenous fate. c, Apln-FlpO;iFlpMTomato-cre/MYfp;Rbpjf/f;Mycf/f mouse survival rate. d, Expression of Apln-HA-FlpO in retina vessels at P6 shows the localization of HA-FlpO in the nuclei of angiogenic front ECs, and not in arteries. e, Schematic of recombination of the Apln-FlpO and iFlpMTomato-cre/MYfp alleles in the retina angiogenic front at P2 and distribution of MbTomato+ and MbYFP+ cells at P6. f–h, Representative confocal images of P6 retinas from mice with the indicated genotypes, and immunostained for isolectinB4, Tomato/Dsred and YFP. Arteries are easily distinguishable from veins by their relatively stronger IsolectinB4 signal and morphology/diameter. i, Quantifications shown are complementary to those in Fig. 4g and allow the direct comparison of significance in the differential distribution of Tomato+ cells only within arteries, or only within capillaries, or only within veins, without taking into account their distinct capillary frequencies and developmental proliferation bias. The results indicate that Rbpj/MycDKO ECs are less frequently found in capillaries, and therefore proliferate less, but have a normal ability of forming arteries, unlike RbpjKO ECs, that occur at a relatively high frequency in capillaries and at very low frequency in arteries (see also log2-transformed fold change indicated in Fig. 4g). j, k, Sectional analysis of control and mutant genetic mosaics of embryonic, newborn and adult hearts showing the very low frequency or absence of Tomato+/mutant cells in coronary arteries of Rbpjflox/flox hearts, and their presence in the coronary arteries of control and Rbpjflox/flox/Mycflox/flox hearts. For quantifications of adult hearts data, see Fig. 4e. l, Sectional analysis of control and mosaic mutant adult livers showing portal arteries (PA) and portal veins (PV) and respective quantifications in these oxygen-rich (DLL4+ and EFNB2+) liver portal vessels. m, Confocal imaging of the inner surface of aortas, stained for CD144 (VE-cadherin), showing the relative occurrence of control and mutant cells and chart with the respective quantifications. Data shown as mean ± s.d. *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars, 100 μm. For statistics, see Supplementary Data 1.
Extended Data Fig. 10 Models for the regulation of arterialization by Notch.
The initial model used to describe arterialization was based on the assumption that an increase in pulsatile and oxygenated blood flow induces the direct differentiation and development of arterial vessels. The current model is based on the evidence that pre-arterial capillary ECs have higher Notch–RBPJ activity and higher expression of arterial genes. This model assumes that arterial fate is determined before the increase in arterial blood flow, through the direct induction of arterial gene expression via the Notch–RBPJ transcriptional complex. Here, we propose a model in which Notch–RBPJ suppresses MYC-dependent cell cycle or biosynthetic activity, rendering ECs more permissive to the adoption of an arterial phenotype without the need for Notch-dependent genetic pre-determination or differentiation. This model is supported by the observed phenotypic and transcriptomic outcomes in several mouse mutant models.
Supplementary information
Supplementary Figure
Supplementary Figure 1: Original Western Blots.
Supplementary Figure
Supplementary Figure 2: FACS data and gatings used to isolate endothelial cells.
Supplementary Data
Supplementary File 1: All statistics for Figures 1–4 and Extended Data Figures 1–10.
Supplementary Table
Supplementary Table 1: Primers used to genotype the different mouse alleles.
Supplementary Table
Supplementary Table 2: Tamoxifen injections and dissection stages.
Supplementary Table
Supplementary Table 3: List of antibodies.
Supplementary Table
Supplementary Table 4: Conditions and stages used to collect ECs by FACS.
Supplementary Table
Supplementary Table 5: List of qRT-PCR Taqman assays.
Source data
Rights and permissions
About this article
Cite this article
Luo, W., Garcia-Gonzalez, I., Fernández-Chacón, M. et al. Arterialization requires the timely suppression of cell growth. Nature 589, 437–441 (2021). https://doi.org/10.1038/s41586-020-3018-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-3018-x
This article is cited by
-
Eph-ephrin signaling couples endothelial cell sorting and arterial specification
Nature Communications (2024)
-
Parenchymal cues define Vegfa-driven venous angiogenesis by activating a sprouting competent venous endothelial subtype
Nature Communications (2024)
-
CD32 captures committed haemogenic endothelial cells during human embryonic development
Nature Cell Biology (2024)
-
Gene regulation for inflammation and inflammation resolution differs between umbilical arterial and venous endothelial cells
Scientific Reports (2023)
-
A Notch between vascular morphogenesis and transcriptional identity
Nature Cardiovascular Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.