Abstract
Compartmentalization of cellular material in droplet-like structures is a hallmark of liquid–liquid phase separation1,2, but the mechanisms of droplet removal are poorly understood. Evidence suggests that droplets can be degraded by autophagy3,4, a highly conserved degradation system in which membrane sheets bend to isolate portions of the cytoplasm within double-membrane autophagosomes5,6,7. Here we examine how autophagosomes sequester droplets that contain the protein p62 (also known as SQSTM1) in living cells, and demonstrate that double-membrane, autophagosome-like vesicles form at the surface of protein-free droplets in vitro through partial wetting. A minimal physical model shows that droplet surface tension supports the formation of membrane sheets. The model also predicts that bending sheets either divide droplets for piecemeal sequestration or sequester entire droplets. We find that autophagosomal sequestration is robust to variations in the droplet-sheet adhesion strength. However, the two sides of partially wetted sheets are exposed to different environments, which can determine the bending direction of autophagosomal sheets. Our discovery of this interplay between the material properties of droplets and membrane sheets enables us to elucidate the mechanisms that underpin droplet autophagy, or ‘fluidophagy’. Furthermore, we uncover a switching mechanism that allows droplets to act as liquid assembly platforms for cytosol-degrading autophagosomes8 or as specific autophagy substrates9,10,11. We propose that droplet-mediated autophagy represents a previously undescribed class of processes that are driven by elastocapillarity, highlighting the importance of wetting in cytosolic organization.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The plasmids and experimental data that support the findings of this study are available from the corresponding author upon reasonable request. Gel raw data are provided as Supplementary Figures. Figures describing the results of the model can be reconstructed directly from the equations and procedures presented in the paper and in the Supplementary Methods, and require no data. Source data are provided with this paper.
Change history
28 January 2021
This article originally published with an incorrect Published Online date of 9 December 2020. This has been amended to the correct Published Online date of 20 January 2021.
References
Shin, Y. & Brangwynne, C. P. Liquid phase condensation in cell physiology and disease. Science 357, eaaf4382 (2017).
Bergeron-Sandoval, L.-P. & Michnick, S. W. Mechanics, structure and function of biopolymer condensates. J. Mol. Biol. 430, 4754–4761 (2018).
Buchan, J. R., Kolaitis, R.-M., Taylor, J. P. & Parker, R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 153, 1461–1474 (2013).
Zhang, G., Wang, Z., Du, Z. & Zhang, H. mTOR regulates phase separation of PGL granules to modulate their autophagic degradation. Cell 174, 1492–1506 (2018).
Knorr, R. L., Dimova, R. & Lipowsky, R. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One 7, e32753 (2012).
Agudo-Canalejo, J. & Knorr, R. L. in Autophagy: Methods and Protocols (eds Ktistakis, N. & Florey, O.) 173–188 (Springer, 2019).
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011).
Fujioka, Y. et al. Phase separation organizes the site of autophagosome formation. Nature 578, 301–305 (2020).
Sun, D., Wu, R., Zheng, J., Li, P. & Yu, L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 28, 405–415 (2018).
Sánchez-Martín, P. & Komatsu, M. p62/SQSTM1 – steering the cell through health and disease. J. Cell Sci. 131, jcs222836 (2018).
Zaffagnini, G. et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 37, e98308 (2018).
de Gennes, P. G. Wetting: statics and dynamics. Rev. Mod. Phys. 57, 827–863 (1985).
Bico, J., Reyssat, É. & Roman, B. Elastocapillarity: when surface tension deforms elastic solids. Annu. Rev. Fluid Mech. 50, 629–659 (2018).
Yamasaki, A. et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol. Cell 77, 1163–1175 (2020).
Alberti, S. & Hyman, A. A. Are aberrant phase transitions a driver of cellular aging? BioEssays 38, 959–968 (2016).
Jawerth, L. M. et al. Salt-dependent rheology and surface tension of protein condensates using optical traps. Phys. Rev. Lett. 121, 258101 (2018).
Liu, Y., Agudo-Canalejo, J., Grafmüller, A., Dimova, R. & Lipowsky, R. Patterns of flexible nanotubes formed by liquid-ordered and liquid-disordered membranes. ACS Nano 10, 463–474 (2016).
Beutel, O., Maraspini, R., Pombo-García, K., Martin-Lemaitre, C. & Honigmann, A. Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 179, 923–936 (2019).
Long, M. S., Jones, C. D., Helfrich, M. R., Mangeney-Slavin, L. K. & Keating, C. D. Dynamic microcompartmentation in synthetic cells. Proc. Natl Acad. Sci. USA 102, 5920–5925 (2005).
Feric, M. et al. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686–1697 (2016).
Haluska, C. K. et al. Combining fluorescence lifetime and polarization microscopy to discriminate phase separated domains in giant unilamellar vesicles. Biophys. J. 95, 5737–5747 (2008).
Walter, H. & Brooks, D. E. Phase separation in cytoplasm, due to macromolecular crowding, is the basis for microcompartmentation. FEBS Lett. 361, 135–139 (1995).
Klein, I. A. et al. Partitioning of cancer therapeutics in nuclear condensates. Science 368, 1386–1392 (2020).
Johansen, T. & Lamark, T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 432, 80–103 (2019).
Wurzer, B. et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife 4, e08941 (2015).
Riback, J. A. et al. Composition-dependent thermodynamics of intracellular phase separation. Nature 581, 209–214 (2020).
Gatica, D., Lahiri, V. & Klionsky, D. J. Cargo recognition and degradation by selective autophagy. Nat. Cell Biol. 20, 233–242 (2018).
Ngugen, N., Shteyn, V. & Melia, T. J. Sensing membrane curvature. in macroautophagy. J. Mol. Biol. 429, 457–472 (2017).
Gracià, R. S., Bezlyepkina, N., Knorr, R. L., Lipowsky, R. & Dimova, R. Effect of cholesterol on the rigidity of saturated and unsaturated membranes: fluctuation and electrodeformation analysis of giant vesicles. Soft Matter 6, 1472–1482 (2010).
Campeau, E. et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529 (2009).
Dull, T. et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72, 8463–8471 (1998).
Pankiv, S. et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145 (2007).
Tsuboyama, K. et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 354, 1036–1041 (2016).
Itakura, E. & Mizushima, N. p62 targeting to the autophagosome formation site requires self-oligomerization but not LC3 binding. J. Cell Biol. 192, 17–27 (2011).
Morita, K. et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 217, 3817–3828 (2018).
Saitoh, T., Nakano, H., Yamamoto, N. & Yamaoka, S. Lymphotoxin-β receptor mediates NEMO-independent NF-κB activation. FEBS Lett. 532, 45–51 (2002).
Chino, H., Hatta, T., Natsume, T. & Mizushima, N. Intrinsically disordered protein TEX264 mediates ER-phagy. Mol. Cell 74, 909–921 (2019).
Kitamura, T. et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31, 1007–1014 (2003).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Wenzel, E. M. et al. Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat. Commun. 9, 2932 (2018).
Zhang, H. et al. RNA controls polyQ protein phase transitions. Mol. Cell 60, 220–230 (2015).
Kremer, J. R., Mastronarde, D. N. & McIntosh, J. R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71–76 (1996).
McArthur, K. et al. BAK/BAX macropores facilitate mitochondrial herniation and mtDNA efflux during apoptosis. Science 359, eaao6047 (2018).
Angelova, M. I. & Dimitrov, D. S. Liposome electroformation. Faraday Discuss. Chem. Soc. 81, 303–311 (1986).
Knorr, R. L., Steinkühler, J. & Dimova, R. Micron-sized domains in quasi single-component giant vesicles. Biochim. Biophys. Acta 1860, 1957–1964 (2018).
Kusumaatmaja, H., Li, Y., Dimova, R. & Lipowsky, R. Intrinsic contact angle of aqueous phases at membranes and vesicles. Phys. Rev. Lett. 103, 238103 (2009).
Knorr, R. L., Lipowsky, R. & Dimova, R. Autophagosome closure requires membrane scission. Autophagy 11, 2134–2137 (2015).
Takahashi, Y. et al. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9, 2855 (2018).
Zhen, Y. et al. ESCRT-mediated phagophore sealing during mitophagy. Autophagy 16, 826–841 (2020).
Acknowledgements
We thank U. Dahl Brinch, M. Smestad, S. Takahashi and other members of the Mizushima laboratory for technical support; and K. Watanabe and H. Ichijo for providing access to the osmometer. For discussions and comments on the manuscript, we thank H. Kusumaatmaja, Y. Ohsumi, W. Antonin, N. N. Noda and T. Johansen. For discussions regarding the in vitro system, R.L.K. thanks Y. Li, Y. Liu, Z. Zhao, R. Dimova and R. Lipowsky. This work was supported by Japan Science and Technology Agency (JST) Exploratory Research for Advanced Technology (ERATO, grant number JPMJER1702) to N.M. J.A.-C. acknowledges the support of the US National Science Foundation under MRSEC grant number DMR-1420620. This work was partly supported by the Research Council of Norway through its Centres of Excellence funding scheme, project number 262652. R.L.K. dedicates this manuscript to H. Dietrich, who was instrumental in the conceptualization of the project, but died at an early stage.
Author information
Authors and Affiliations
Contributions
R.L.K. conceived, guided and supervised the project; J.A.-C. developed the theory; S.W.S., H.C., S.M.M., C.S. and I.K.-H. designed and performed live-cell experiments including correlative light and electron microscopy with contributions from N.M., A.B. and H.S.; R.L.K. designed and performed in vitro studies; and R.L.K. and A.I.M. wrote the manuscript with contributions from J.A.-C. and S.W.S. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 LC3-positive membranes associate with and deform p62 droplets.
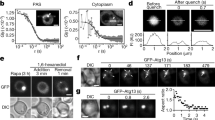
a, p62 droplet contours were obtained by live-cell imaging of RPE1 cells expressing mCherry–p62 and analysed. Binned droplet circularities are plotted as a histogram (n = 421 droplets assessed from three independent experiments). The frequency peak centred at a circularity of 0.90 (Lorentz fit ± s.d., a circularity of 1 indicates a perfect circle). b, Non-circularity correlates with increasing droplet size. Filled data points represent n > 5 droplets and the open data point n = 1 droplet. Data are from a. c, Four representative droplets were observed continuously for >10 min at 12 frames/min and circularities determined. Circularity is plotted for each single droplet individually over time. Frames in which another droplet moved into the field of view were excluded from analysis. d, Mean circularities of single droplets shown in c; n = data points/droplet (black n = 145, red n = 143, blue n = 136, green n = 139). The mean of the four droplets was 0.91 (s.d. = 0.01). e, f, Multiple small autophagosomes form at the surface of large p62-containing droplets. Representative confocal images of a live cell in Eagle’s balanced salt solution (e) and a fixed specimen prepared for ultrastructural analysis of the structure highlighted in f. All images are of RPE1 cells expressing GFP–LC3 and mCherry–p62. Magnified areas in f show single confocal z-sections of autophagosomal membranes (ii and iii); corresponding tomographic sections are shown in Fig. 1f, g. g, Electron tomography reconstruction of autophagosomal membranes i, ii and iii without (left) or with (right) the p62 droplet. The mean radius of the autophagosomal structures i, ii)and iii was 0.23% ± 0.09% ( ± s.d.) of the droplet radius. h, Schematic of droplet deformation by autophagosomal membranes during piecemeal sequestration, as observed in Fig. 1d, e. i, Immunolabelling of a 200-nm section of RPE1 cells expressing mCherry–p62 with an anti-p62-antibody and 10 nm gold particles. Left, transmission electron microscopy image of the section; right, surface tomography slice. The broken pink line delineates the droplet boundary. j, Left, enlarged surface tomography slice of the region indicated in i (right panel). Right, enlarged tomography slice through the section centre of the same region. Five droplets of the same preparation were examined with similar results (i, j). k, FRAP of proteins localizing to p62 droplets. LC3, p62 and p62(ΔLIR) all localized to droplets. Regions of radius a = 1.7 μm were bleached and the recovery of fluorescence recorded. The normalized intensity change over time was fitted to an exponential curve f(x) = A × (1 – e−x/τ) to obtain the apparent recovery time τ and estimate apparent diffusion coefficients (Dapp = a2/τ): DLC3 = 8.44 ± 0.50 μm2/min, Dp62 = 0.54 ± 0.01 μm2/min, and Dp62(ΔLIR) = 0.50 ± 0.01 μm2/min. The curves show mean recovery ± s.d. from 74 ROIs examined over 5 independent experiments (p62), 68 ROIs examined over 3 independent experiments (p62(ΔLIR)) and 62 ROIs examined over 5 independent experiments (LC3). l, Left, FRAP of a droplet-bound autophagosome in RPE1 cells expressing mCherry–p62 and GFP–LC3 (bleach spot and autophagosome as indicated). The green arrow indicates the position of the droplet-attached autophagosome. Intensities of single-channel microscopy images are inverted. Right, line profiles as indicated. Scale bars, 5 μm (e, f), 1 μm (l) or 0.5 μm (i, j). Representative images of n independent experiments (n = 2 for f, l; n = 7 for e).
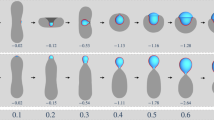
Extended Data Fig. 2 Modelling of droplet–membrane-sheet interaction unveils wetting as a determinant of droplet autophagy.
a, Contact angles of sheets at flat droplet surfaces. There are three distinct wetting states depending on the value of the contact angle. We focus on partial wetting (0° < θin < 180°) throughout this study. Sheets in contact with homogeneous, non-phase-separated solutions correspond to dewetting and complete wetting, as described previously5. For pinned shapes as depicted here (when the three-phase contact line is located at the rim), the interfacial energy of the partial wetting states is roughly independent of the specific value of the contact angle, because for thin sheets with rrim ≪ rsh the area of the rim is negligible with respect to the area of the rest of the sheet. Meanwhile, the value of θin becomes important for unpinned sheets (see b–e, g, h). Instability of the open sheet state (which is a pinned shape) towards the closure of the autophagosome is the only state relevant to this study. Therefore the stability diagrams in f and in Figs. 2d, e, 4a, Extended Data Fig. 5a, b are independent of the value of θin. b–e, Energy landscapes for autophagosome formation. Msh = 1/Rsh is the mean curvature of the sheet. Mshrsh = 2 corresponds to piecemeal autophagy of the droplet, Mshrsh = −2 to cytosolic autophagy, and the vertical dotted lines correspond to Msh = 1/Rdrop; that is, the state in which droplet and sheet curvatures coincide and the sheet wets the droplet without deforming it. The three coloured lines (in b–e) correspond to different values of rsh/rrim (in b, c) or \({\Sigma }_{{\rm{cd}}}{r}_{{\rm{rim}}}^{2}/\kappa \) (in d, e). In all cases, the energy baseline is arbitrary, and the curves have been shifted vertically for clarity to prevent overlapping. In b, c, the sheet grows (blue–yellow–red) at a constant droplet surface tension of \({\Sigma }_{{\rm{cd}}}{r}_{{\rm{rim}}}^{2}/\kappa \)= 0.04. In d, e, the surface tension decreases (blue–yellow–red) at a constant sheet size of rsh/rrim = 20. The Msh = 1/Rdrop state is typically stable for small sheet size or high surface tension (blue lines). As the sheet size increases or the surface tension decreases, this state eventually becomes marginally stable (yellow lines; these lines correspond to the values of Σcd and rsh at which the energy barrier disappears and were used to develop the stability diagrams shown in f and in Figs. 2d, e, 4a, Extended Data Fig. 5a, b) and finally unstable (red lines). The instability occurs towards piecemeal autophagy in b, d, which correspond to zero spontaneous curvature contrast (mcd = 0), or towards cytosolic autophagy in c, e, which correspond to negative contrast (mcdRdrop = −3). In all cases, the solid lines correspond to θin = π/4, and the dashed lines to θin = 3π/4. Both lines overlap over a wide region at intermediate values of Msh, which correspond to the pinned state, and diverge in the unpinned state. Because the instability occurs within the pinned region, it is independent of the value of θin. Specific values used are (given in the order blue–yellow–red): b rsh/rrim = 5, 12, 20; c rsh/rrim = 5, 8.3, 20; d \({\Sigma }_{{\rm{cd}}}{r}_{{\rm{rim}}}^{2}/\kappa \) = 0.2, 0.068, 0.02; and e \({\Sigma }_{{\rm{cd}}}{r}_{{\rm{rim}}}^{2}/\kappa \)= 0.2, 0.054, 0.02. Landscapes can be compared with the stability diagrams b1 and b4 in Extended Data Fig. 5b. The droplet radius is set to Rdrop = 50rrim in all cases. f, Stability diagrams (as shown in Fig. 2d, e) for droplets of sizes Rdrop/rrim = 10, 20, 40, 80 that have a corresponding droplet radius of 0.25, 0.5, 1 or 2 μm (for sheets with rrim = 25 nm, as in Fig. 2). For each droplet size, we also display a horizontal dotted line for rsh = 2Rdrop that shows the sheet size required for complete droplet sequestration, which increases strongly with droplet size. These lines demonstrate that complete sequestration of large droplets is unlikely, owing to the very large sheet required. Meanwhile, the minimal surface tension necessary for complete droplet engulfment is smaller for larger droplets. g, h, Unpinning of the sheet rim from the droplet surface. Four sequences demonstrating morphological changes of sheets developing at droplets. Two contact angles (45° in g, 135° in h) and both directions of bending, either enclosing the droplet (top panels) or the cytosol (bottom panels) are portrayed in these scenarios. All shapes are calculated including the correct volume, area and contact angle constraints, for Rdrop/rsh = 0.82. The two sides of the sheet (green) cannot be resolved owing to their close proximity. Thus, the sheets are depicted as a single line. Vertical lines indicate transitions between pinned and unpinned shapes.
Extended Data Fig. 3 Formation of membrane sheets observed in vivo and in vitro.
a, Live-cell images of droplet autophagy in RPE1 cells expressing GFP–LC3 and mCherry–p62. Autophagosome formation resulted in droplet division. b, Time series of the piecemeal process highlighted in the boxed regions (a) and shown in Fig. 2f. c, Several examples of droplet autophagy whereby two distinct droplets form. Small droplets (circled) with a diameter of 1.0 ± 0.15 μm (mean ± s.d., n = 4 sheets on distinct droplets) were sequestered within LC3-positive autophagosomes, with large droplets essentially free of remaining autophagosomal membranes. Duration of droplet splitting: 10.6 min ± 6.1 min (mean ± s.d.). d, Expansion of LC3 sheets. t = 0 min corresponds to the maximum sheet length. Rate = 0.2 ± 0.04 μm/min. Maximal length = 2.7 μm ± 0.7 μm. Duration 10.1 min ± 3.3 min. Droplets with two intensity peaks occurred within an additional 4.1 min ± 2.6 min (mean ± s.d.). n = 4 sheets on distinct droplets shown in b, c. e, Representative GUVs before phase separation. Interior tubules that form by PEG adsorption onto GUV membranes distribute randomly within the homogeneous interior solution. A fluorescent phospholipid (Atto633–DOPE) and polymer (FITC–dextran 500 kDa) were used to label the membrane (green) and internal solution (magenta). f, g, Representative GUV after phase separation. Vertical confocal (xz) section (f) and horizontal confocal (xy) sections (g) at the droplet surface of this GUV highlight the tubular membrane network that wets the droplet surface. Tubule formation and wetting tubules (e–g) confirm previous work17. h, Tubules visualized by a water-soluble dye (yellow; 5 μM sulforhodamine B) added externally to the GUV. i, The tubular network shown in g after an additional 4-min incubation. The de novo formation of two circular membrane structures representing membrane sheets is observed (arrows). j, Quantification of the relative abundance of membrane shapes at the interface after hyperosmotic deflation (1 h) and following an additional 16-h equilibration (three independent experiments with more than 50 GUVs each, mean ± s.d. with single data points). k, Representative examples of tubules and sheets at droplet surfaces (single xy sections at the droplet surface; the central image is a maximum intensity projection of two xy sections). Scale bars, 2 μm (b, c) and 5 μm (all other images). Representative images of n independent experiments (n = 5 (a–c); n = 10 (e–g, i, k) and n = 4 (h)).
Extended Data Fig. 4 Membrane sheets close to form double-membrane vesicles in vitro.
a, Fluorescence intensity line profiles of Sh1 and Sh2 (as indicated in Fig. 3b). The profiles intersect the two sides of the sheet membrane and the single bilayer of the GUV, as labelled. b, Luminal staining of sheets by addition of the water-soluble, membrane-impermeable dye sulforhodamine B outside GUVs (yellow). The orange line indicates the position of the intensity profile. c, Remodelling of a droplet-pinned, predominantly tubular structure stained by sulforhodamine B into a circular membrane sheet, as observed by confocal time-lapse imaging (inverted intensity). Top panels, droplet surface imaged before (left) and after (right) remodelling. Bottom panels, time series of remodelling. d, The size of sheets can be stable for extended periods. Line profiles of Sh1 and Sh2 obtained from xy sections shown in left panels of Fig. 3b, c. e, Membranes wet droplet surfaces in a GUV-free assay. Droplets sedimenting on a pre-hydrated membrane were observed by confocal microscopy. Maximum intensity projections of distinct upper droplet hemispheres showing surface pinning of membranes in the form of (from left to right) small vesicles, narrow tubules, wide tubules while remodelling into a sheet, or two sheets with small vesicles. f, A representative vertical confocal section showing the profile of a sedimented droplet with a surface-attached, droplet-sequestering membrane sheet (left). The corresponding maximum intensity projection is shown at right. g, In some cases, both sides of wetting double-membrane sheets are resolvable. Left, a single confocal section with an orange line indicating the fluorescence intensity profile. Centre, line profile exhibiting a membrane double peak. Right, maximum intensity projection of the same droplet. h, Left, confocal section of the centre plane of a droplet undergoing sequestration. Also shown are maximum intensity projections of the upper (centre) and lower (right) droplet hemispheres. i, Fluorescence intensity line profile of a double membrane vesicle and the preceding sheet, Sh3 (shown in Fig. 3b, c). The size of the structure decreased by approximately half during remodelling. A fluorescent phospholipid (Atto633–DOPE) and polymer (FITC-dextran 500 kDa) were used to label the membrane (green) and droplet (magenta). j, Luminal staining of a double membrane vesicle by addition of the water-soluble, membrane-impermeable dye sulforhodamine B outside GUVs (yellow). The orange line indicates the intensity profile segment. k, Multiple GUVs with numerous wetting sheets are shown under isotonic conditions (left) and 7 min after exposure to hypoosmotic conditions (right). The sheets are partially out-of-focus because the z position of the slightly non-horizontal droplet surface differs between GUVs. A few minutes after the decrease in Σcd associated with hypoosmotic conditions, all sheets closed into autophagosome-like double-membrane vesicles. Confocal fluorescence microscopy of Atto633–DOPE-stained membranes (intensities inverted). l, Intensity plots for segments intersecting double-membrane vesicles and corresponding GUVs as indicated in k (right). m, Left, membrane fluorescence intensity of GUVs delimiting membranes and their internal double-membrane vesicles. Gradient determined by linear regression (n = 9 GUVs examined, several double-membrane vesicles within one GUV are shown as mean ± s.d.). Right, size measurements of double-membrane vesicles relative to preceding sheets as illustrated in i. Gradient determined by linear regression (n = 13 sheet-vesicle pairs). Scale bars, 20 μm (k) and 5 μm (all other images). Representative results from n independent experiments (n = 5 (k–m), n = 3 (c) and n = 2 (b, e–h, j).
Extended Data Fig. 5 Membrane asymmetry controls sheet bending direction.
a, A decrease in droplet surface tension triggers sheet closure into double-membrane vesicles. Morphologies predicted by the physical model under conditions of decreasing surface tension as a function of the spontaneous curvature asymmetry between both sides of the membrane sheet, mcd, and the sheet radius, rsh. The droplet radius is set to Rdrop = 50rrim. b, Stability diagrams as a function of spontaneous curvature contrast, mcd. Stability diagrams in the (Σcd, rsh) plane for b1: mcd = −3/Rdrop, b2: mcd = −2/Rdrop, b3: mcd = −1/Rdrop, b4: mcd = 0 and b5: mcd = +2/Rdrop. In all cases, the droplet radius is set to Rdrop = 50rrim, and white, blue, and yellow regions signify wetting of the open sheet (shape A), piecemeal autophagy (shape B) and cytosolic autophagy (shape E), respectively, as indicated in b2. c, Double-membrane autophagosome-like vesicles (AV) sequester both liquid phases present in GUVs. The formation of AV1, AV2 and AV3, AV4 was observed in GUV1 and GUV2, respectively, at phase-separated droplet surfaces within the GUV. d, The quantified relative mean fluorescence intensity of sequestered volumes shown in c indicate that AV1 and AV2 sequestered the droplet, whereas AV3 and AV4 sequestered the bulk (non-droplet) interior. For normalization, the intensity of the droplet was set to 1 and the exterior intensity to zero. Representative results from three independent experiments. e, Schematic of the AV geometries identified in c, d. f, p62 droplets that lack the LIR domain retain liquid-like behaviour of p62 droplets. p62(ΔLIR) droplet circularities were determined, binned and plotted as a histogram (n = 399 droplets, observed across three independent experiments). Circularity peaks at 0.91 (Lorentz fit ± s.d.), a similar result to that for p62 (Extended Data Fig. 1a–d). g, Correlation between droplet circularity and size for the dataset shown in f. Filled data points correspond to n > 5 droplets, the open data point to n = 1 droplet. h, Droplets containing p62(ΔLIR) coalesce and show the shape relaxation characteristic of condensates with liquid-like properties. Snapshots of a single coalescence event (top panels; numbers indicate time in min). Shape relaxation of the droplet shown fitted by exponential regression. Relaxation time = 0.93 ± 0.14 min (s.d.). i, Relaxation times ± s.d. of multiple coalescing p62(ΔLIR) droplets obtained from fitting, an in h. Linear fit slope 0.79 min/μm ± 0.58 min/μm (n = 8 droplets examined over three independent experiments). Experiments were performed in RPE1 cells expressing mCherry–p62(ΔLIR). Fluorescence intensities are inverted. Scale bars, 5 μm (c) or 2 μm (h).
Extended Data Fig. 6 Autophagosomal membranes wet and sequester p62 droplets in a physiological context.
a, Confocal images of fixed RPE1 cells stably expressing GFP–LC3 and mCherry–p62 or mCherry–p62(ΔLIR). Cytosolic p62 droplet formation (droplets were not found within LAPM1-positive lysosomes or engulfed by LC3-positive vesicles) was induced by addition of the reversible VPS34 inhibitor SAR405 (10 μM) for 4 h. Arrows in the left panels indicate magnified panels in the right panels. b, Droplet formation of p62 mutants unable to bind ubiquitin (M404V) or to self-oligomerize via the PB1 domain (R21A) is inhibited. Indicated cells were treated with SAR405 for 4 h. Corresponding wild-type and ΔLIR data are shown in Extended Data Fig. 7b. c, Western blots of total cell lysates of each stable RPE1 cell line expressing GFP–LC3 and an siRNA-resistant mCherry–p62 variant (wild type (WT), ΔLIR, M404V or R21A) with or without endogenous p62 knockdown using siRNA. d, Left, knockdown efficiency of siRNA treatment. Right, quantification of mCherry–p62 expression levels (WT, ΔLIR, M404V or R21A) relative to endogenous p62. Data are mean ± s.d. from three independent experiments. e, f, RPE1 cells treated with full medium (DMEM/F12) after 4-h VPS34 inhibition by SAR405 and subsequent formation of cytosolic p62 droplets. GFP–LC3 localized to the surface of p62 (e) and p62(ΔLIR) (f) droplets within 15 min of SAR405 washout. Arrows in the top panels indicate magnified panels in the bottom panels. g, Piecemeal sequestration of a p62 droplet by GFP–LC3 after SAR405 washout. Live-cell imaging of droplets was initiated 15 min after SAR405 washout and z-stacks of a whole cell were acquired every 15 s. 0 min was defined as the time point at which the GFP–LC3 signal exhibited maximum length. Representative images from two (a, b, e, f, g) or three (c, d) independent experiments. Scale bars, 10 μm (a, b), 5 μm (e, f) or 1 μm (g, magnified images in a, e, f).
Extended Data Fig. 7 Disruption of the LIR region perturbs droplet autophagy.
a, Localization of endogenous ubiquitin and siRNA-resistant mCherry–p62 (left) or siRNA-resistant mCherry–p62(ΔLIR) (right) in RPE1 cells. The mCherry–p62(ΔLIR) lacks amino acids 321–348 that make up the LIR. Stable p62 expression is quantified in Extended Data Fig. 6d. b, Left, autophagy was inhibited by SAR405 treatment for 4 h before release by DMEM washout (additional data in Extended Data Fig. 6b). Right, droplet numbers per cell were determined by ubiquitin immunofluorescence before and after 4h SAR405 treatment, and also following SAR405 washout. Exponential fits (lines) yield time constants ± s.d. of 37.03 ± 10.39 min (ΔLIR), 19.25 ± 1.17 min (WT), 17.24 ± 0.02 min (M404V) and 16.14 ± 6.94 min (R21A) that characterize droplet turnover and are plotted as inset. n values, given as fields of view (and cell numbers) at −240, 0, 30, 60 and 120 min after SAR405 treatment: WT, n = 6 (118), 5 (127), 5 (175), 6 (178), 7 (154); ΔLIR, n = 10 (120), 5 (109), 9 (206), 7 (169), 6 (172); M404V, n = 6 (127), 5 (129), 7 (171), 5 (158), 5 (123); R21A, n = 5 (125), 4 (135), 6 (163), 5 (118), 5 (135). Data are mean ± s.d. c, The region within the white broken boundary (left) was examined by electron tomography, showing that the droplet is not sequestered by LC3-positive membranes (arrows). These data correspond to the image presented in Fig. 4b. d, Time series (30-s interval) of a wild-type MEF exhibiting a WIPI2-positive structure sequestering a subset of a p62 droplet. Droplet deformation and division are highlighted by arrows (wild-type MEF expressing WIPI2–mRuby3 and GFP–p62). e, f, Correlated confocal (e) and scanning electron (f) microscopy of autophagosomes attached to the p62 droplet indicated in the leftmost panel. The regions indicated by numbers are shown at high magnification in the rightmost panels of f. The autophagosome interior is filled with low-density material consistent with sequestration of droplet material, not ribosomes present in the cytosol (MEF expressing WIPI2–mRuby3 and GFP–p62). g, Time series (at 30-s intervals) of an ATG3(KO) MEF in which a droplet-bound WIPI2-positive structure is observed to expand, forming a cup-shape intermediate before closing without droplet sequestration (ATG3(KO) MEF expressing WIPI2–mRuby3 and GFP–p62). h, Several additional examples of droplet-bound WIPI2-positive structures that expand towards the cytosol, subsequently closing to form autophagosomes without droplet sequestration (circles), as quantified in Fig. 4c (ATG3(KO) MEFs expressing WIPI2–mRuby3 and GFP–p62). Scale bars for fluorescence images, 2 μm; scale bar for electron microscopy, 0.5 μm (c, left panel in f), 0.2 μm (right panels in f). Representative images from three (d, g, h) or two (c, e, f) independent experiments. i, Size of p62 droplets increases in ATG3(KO) cells. Quantification of the diameter of p62 droplets in wild-type and ATG3(KO) MEFs. Data medians are shown within boxes representing the interquartile range (25th to 75th percentile), with whiskers denoting the 10th and 90th percentiles. Significance was determined by the unpaired two-tailed Student’s t-test (P = 6.2 × 10−10). Data were collected in three independent experiments and n ≥ 100 p62 droplets were observed for each cell type (control n = 105, ATG3(KO) n = 100).
Extended Data Fig. 8 Model of droplet autophagy and parameters used for calculations.
a, The LIR interaction controls bending direction during droplet autophagy (Fig. 4, Extended Data Fig. 7). Two coexisting liquids are characterized by dissimilar physicochemical properties and molecule concentrations. This implies that the spontaneous curvatures of membranes in contact with either the cytosol or the droplet will be different (\({m}_{{\rm{c}}}\ne {m}_{{\rm{d}}}\)). Thus, symmetric sheets fully immersed in homogeneous liquids (corresponding to dewetting and complete wetting in this manuscript, Extended Data Fig. 2a) have an equal probability of bending in both directions because a spontaneous curvature contrast between both sheet sides does not exist. Any membrane asymmetry introduced (indicated by the blue line), for example, by membrane-binding proteins and complexes, can induce a preferred bending direction by generating a spontaneous curvature contrast5. During droplet autophagy, both sheet sides contact two different liquids (the cytosol and the droplet). Thus, it is very unlikely that the spontaneous curvatures of both sides of a sheet are equal, that is, mcd = 0. In general, a wetting sheet can therefore be considered asymmetric, that is, \({m}_{{\rm{cd}}}\ne 0\). b, Geometric parameters for numerical calculations. Pinned shapes are determined by the sheet curvature radius Rsh and angle θsh, and the droplet curvature radius Rd and angle θd. To determine unpinned shapes, the angle θco, which defines the location of the three-phase contact line, is also required. For unpinned shapes, the intrinsic contact angle θin (between the tangents of the two circles) imposes a relation between θco and θd, as given by Supplementary equation (12) in the Supplementary Methods for the two possible (+) shapes, and by Supplementary equation (13) in the Supplementary Methods for the (−) shape.
Supplementary information
Supplementary Information
This PDF file includes Supplementary Methods (This file presents all equations and procedures to reconstruct our theoretical model.). Supplementary Figures 1-3 (These Figures show gel raw data.).
Rights and permissions
About this article
Cite this article
Agudo-Canalejo, J., Schultz, S.W., Chino, H. et al. Wetting regulates autophagy of phase-separated compartments and the cytosol. Nature 591, 142–146 (2021). https://doi.org/10.1038/s41586-020-2992-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2992-3
This article is cited by
-
Membrane transformations of fusion and budding
Nature Communications (2024)
-
Kinetic control of shape deformations and membrane phase separation inside giant vesicles
Nature Chemistry (2024)
-
Sequence-dependent material properties of biomolecular condensates and their relation to dilute phase conformations
Nature Communications (2024)
-
The mechanisms and roles of selective autophagy in mammals
Nature Reviews Molecular Cell Biology (2023)
-
How to drink like a liposome
Nature Reviews Chemistry (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.