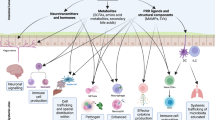
Abstract
The liver connects the intestinal portal vasculature with the general circulation, using a diverse array of immune cells to protect from pathogens that translocate from the gut1. In liver lobules, blood flows from portal triads that are situated in periportal lobular regions to the central vein via a polarized sinusoidal network. Despite this asymmetry, resident immune cells in the liver are considered to be broadly dispersed across the lobule. This differs from lymphoid organs, in which immune cells adopt spatially biased positions to promote effective host defence2,3. Here we used quantitative multiplex imaging, genetic perturbations, transcriptomics, infection-based assays and mathematical modelling to reassess the relationship between the localization of immune cells in the liver and host protection. We found that myeloid and lymphoid resident immune cells concentrate around periportal regions. This asymmetric localization was not developmentally controlled, but resulted from sustained MYD88-dependent signalling induced by commensal bacteria in liver sinusoidal endothelial cells, which in turn regulated the composition of the pericellular matrix involved in the formation of chemokine gradients. In vivo experiments and modelling showed that this immune spatial polarization was more efficient than a uniform distribution in protecting against systemic bacterial dissemination. Together, these data reveal that liver sinusoidal endothelial cells sense the microbiome, actively orchestrating the localization of immune cells, to optimize host defence.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus database with the accession number GSE144087. Source data are provided with this paper.
Code availability
The modelling code can be accessed via GitHub at https://github.com/UniboDIFABiophysics/LiverBacteriaCaptureAssay.
Change history
09 August 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-03346-0
24 October 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41586-022-05462-x
References
Kubes, P. & Jenne, C. Immune responses in the liver. Annu. Rev. Immunol. 36, 247–277 (2018).
Kastenmüller, W., Torabi-Parizi, P., Subramanian, N., Lämmermann, T. & Germain, R. N. A spatially-organized multicellular innate immune response in lymph nodes limits systemic pathogen spread. Cell 150, 1235–1248 (2012).
Baptista, A. P. et al. The chemoattractant receptor Ebi2 drives intranodal naive CD4+ T cell peripheralization to promote effective adaptive immunity. Immunity 50, 1188–1201 (2019).
Doi, Y. et al. Development of complementary expression patterns of E- and N-cadherin in the mouse liver. Hepatol. Res. 37, 230–237 (2007).
Braeuning, A. et al. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 273, 5051–5061 (2006).
Bouwens, L., Baekeland, M., De Zanger, R. & Wisse, E. Quantitation, tissue distribution and proliferation kinetics of Kupffer cells in normal rat liver. Hepatology 6, 718–722 (1986).
Sleyster, E. C. & Knook, D. L. Relation between localization and function of rat liver Kupffer cells. Lab. Invest. 47, 484–490 (1982).
Itoh, Y. et al. Functional heterogeneity of rat liver macrophages: interleukin-1 secretion and Ia antigen expression in contrast with phagocytic activity. Liver 12, 26–33 (1992).
Gerner, M. Y., Kastenmuller, W., Ifrim, I., Kabat, J. & Germain, R. N. Histo-cytometry: a method for highly multiplex quantitative tissue imaging analysis applied to dendritic cell subset microanatomy in lymph nodes. Immunity 37, 364–376 (2012).
MacParland, S. A. et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 9, 4383 (2018).
Geissmann, F. et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 3, e113 (2005).
Benhamouche, S. et al. Apc tumor suppressor gene is the “zonation-keeper” of mouse liver. Dev. Cell 10, 759–770 (2006).
Burke, Z. D. et al. Liver zonation occurs through a beta-catenin-dependent, c-Myc-independent mechanism. Gastroenterology 136, 2316–2324 (2009).
Shiojiri, N. et al. Heterogeneous hepatocellular expression of glutamine synthetase in developing mouse liver and in testicular transplants of fetal liver. Lab. Invest. 72, 740–747 (1995).
Schloss, P. D. et al. Stabilization of the murine gut microbiome following weaning. Gut Microbes 3, 383–393 (2012).
Mao, K. et al. Innate and adaptive lymphocytes sequentially shape the gut microbiota and lipid metabolism. Nature 554, 255–259 (2018).
Jacob, A. I., Goldberg, P. K., Bloom, N., Degenshein, G. A. & Kozinn, P. J. Endotoxin and bacteria in portal blood. Gastroenterology 72, 1268–1270 (1977).
Lynch, R. W. et al. An efficient method to isolate Kupffer cells eliminating endothelial cell contamination and selective bias. J. Leukoc. Biol. 104, 579–586 (2018).
Greenhalgh, S. N., Conroy, K. P. & Henderson, N. C. Cre-ativity in the liver: transgenic approaches to targeting hepatic nonparenchymal cells. Hepatology 61, 2091–2099 (2015).
Weber, M. et al. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 339, 328–332 (2013).
Halpern, K. B. et al. Paired-cell sequencing enables spatial gene expression mapping of liver endothelial cells. Nat. Biotechnol. 36, 962–970 (2018).
García-López, M. A. et al. CXCR3 chemokine receptor distribution in normal and inflamed tissues: expression on activated lymphocytes, endothelial cells, and dendritic cells. Lab. Invest. 81, 409–418 (2001).
Thomas, S. Y. et al. CD1d-restricted NKT cells express a chemokine receptor profile indicative of Th1-type inflammatory homing cells. J. Immunol. 171, 2571–2580 (2003).
Groom, J. R. et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity 37, 1091–1103 (2012).
Sakai, M. et al. Liver-derived signals sequentially reprogram myeloid enhancers to initiate and maintain Kupffer cell identity. Immunity 51, 655–670 (2019).
Metzemaekers, M., Vanheule, V., Janssens, R., Struyf, S. & Proost, P. Overview of the mechanisms that may contribute to the non-redundant activities of interferon-inducible CXC chemokine receptor 3 ligands. Front. Immunol. 8, 1970 (2018).
Vanheule, V. et al. CXCL9-derived peptides differentially inhibit neutrophil migration in vivo through interference with glycosaminoglycan interactions. Front. Immunol. 8, 530 (2017).
Lämmermann, T. et al. Neutrophil swarms require LTB4 and integrins at sites of cell death in vivo. Nature 498, 371–375 (2013).
Fernandez-Ruiz, D. et al. Liver-resident memory CD8+ T cells form a front-line defense against malaria liver-stage infection. Immunity 45, 889–902 (2016).
Gola, A. et al. Prime and target immunization protects against liver-stage malaria in mice. Sci. Transl. Med. 10, eaap9128 (2018).
Sörensen, I., Adams, R. H. & Gossler, A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood 113, 5680–5688 (2009).
Li, W., Germain, R. N. & Gerner, M. Y. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl Acad. Sci. USA 114, E7321–E7330 (2017).
Goddard, E. T., Fischer, J. & Schedin, P. A portal vein injection model to study liver metastasis of breast cancer. J. Vis. Exp. 118, 54903 (2016).
Pentecost, M., Kumaran, J., Ghosh, P. & Amieva, M. R. Listeria monocytogenes internalin B activates junctional endocytosis to accelerate intestinal invasion. PLoS Pathog. 6, e1000900 (2010).
Heymann, F. et al. Long term intravital multiphoton microscopy imaging of immune cells in healthy and diseased liver using CXCR6.Gfp reporter mice. J. Vis. Exp. 97, 52607 (2015).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
McCarthy, D. J., Chen, Y. & Smyth, G. K. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297 (2012).
Sergushichev, A. A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. Preprint at https://doi.org/10.1101/060012 (2016).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Turner, A. B. R. spatstat: an R Package for analyzing spatial point patterns. J. Stat. Softw. 12, 1–42 (2005).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer-Verlag, 2016).
Acknowledgements
We thank A. Luster and J. Lian for providing REX3 mice; E. Miao for providing (ΔintAB inlAm) Listeria stocks; P. Brook for preliminary versions of the in silico bacterial capture model; Y. Belkaid and N. Bouladoux for GF mice; R. H. Adams for iCdh5-Cre mice; A. Martins for help with the RNA sequencing experiments; C. Chu for critical reading of the manuscript; and all members of the Laboratory of Immune Systems Biology for their comments during the course of these studies and input during preparation of the manuscript. A.P.B. is supported by a Marie Sklodowska-Curie Action (MSCA) fellowship (grant agreement 898090). This research was supported by the Intramural Research Program of the NIAID (NIH).
Author information
Authors and Affiliations
Contributions
A.G. conceptualized, designed and conducted most of the experiments, performed data analysis and prepared the manuscript. M.G.D. and I.D.C.F. helped with the in vitro and in vivo bacterial experiments. E.S. helped with RNA sequencing library preparation and performed RNA sequencing data analysis. C.S. and G.C. generated the in silico bacterial capture model. R.M.S. and A.J.R. performed the analysis on human samples, IBEX imaging and cleared liver sections. H.S.W. and A.P.B. provided discussion, data interpretation and help with statistical analyses. J.M.H. obtained human liver biopsies. R.N.G. designed the experiments, interpreted the data and prepared the manuscript. All authors helped to write and edit the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Matteo Iannacone, Shalev Itzkovitz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Kupffer cell MHCII expression and localization in mouse and human livers.
a, Representative IF image of F4/80+, MHCII, Collagen IV, and E-cadherin expression in mouse liver lobule. (i-iii) Channel shown labelled in figures, CV highlighted by dashed circles in IFs. b, Scatter plot of KC MHCII Mean Fluorescence Intensity (MFI) and distance to the centre of CV vasculature (μm) (n = 3 mice, 2 lobules/mouse, each dot represents a KC). Linear regression model shown, intercept (m), p-value (Two-Tailed t-test) and R2 indicated in figure. c, Representative IF human liver image of Kupffer cells (stained by CD163 and HLA-DR) and liver enzymes ASS1 and GS to identify the liver lobule (CV highlighted by dashed circles in IF images). d, Total number of KCs cells in PP and CV regions normalized to volume, each dot represents an ROI; Median shown. Two-tailed Mann–Whitney; *P = 0.0448. (n = 7 donors). Channels and CV as labelled in image.
Extended Data Fig. 2 NKT cells show periportal enrichment, and are disrupted in iCdh5-Myd88−/− and Cxcl9−/− mice.
a, Representative IF image with CXCR6-GFP (NKT), E-cadherin, and CD1d, CV highlighted by dashed circles in IF images. b, Total number of NKT cells in PP and CV regions normalized to volume, each dot represents an ROI. n = 4 mice, Two-tailed Mann–Whitney, ****P < 0.0001; Median shown. c, Representative image from 2P-intravital movie of CXCR6-GFP animal showing E-Cadherin antibody injected intravenously i.v. and NKT cell positioning at time zero; Bottom: compiled NKT cell tracks. d, Representative ratio of total NKT cell numbers in CV to PP ROIs per frame over time. e, Total number of NKT cells in PP and CV regions normalized to volume for 2-P intravital movies, each dot represents an ROI. n = 3 mice, Two-tailed Mann–Whitney, ****P < 0.0001; Median shown. f, Ratio of total NKT cell numbers in CV to PP ROIs for 2-P intravital movies, each dot represents a frame. n = 3 mice. g, Representative IF image CXCR6+ CD3+ cells (NKT cells), and E-cadherin in iCdh5-MyD88wt/wt, iCdh5-MyD88fl/fl and CXCL9−/− animals. h, i, Ratio of total NKT cells in PP and CV regions normalized to volume (h), and total number of cells per volume (i); each dot represents a lobule. n = 4 mice/condition, Kruskal–Wallis test, Dunn’s multiple comparison test, ***P = 0.0002, **P = 0.0013, ns (not significant) P > 0.9999; Median and quartile range shown. Channels and CV as labelled in image.
Extended Data Fig. 3 Immunofluorescent images and total numbers of Kupffer cells in mice with distinct commensal microbiomes.
a, Total number of KCs normalized to volume in AlbCre-βcateninwt/wt and AlbCre-βcateninfl/fl animals, each dot represents a biological replicate, n = 3 mice/condition, Two-tailed Mann–Whitney test, ns p-value = 0.400; Median shown. b, Representative IF images of mouse livers at days (D) 3, 8, 13, 20, and 25 post-partum. At D3, GS synthetase expression is absent and E-cadherin is homogenously expressed throughout the liver lobule; by D8 both molecules show zonation. c, Same representative IF images of mouse livers at days (D) 3, 8, 13, 20, and 25 post-partum showing E-cadherin and F4/80+ KCs (for quantification, see Fig. 1h). d, Total number of KCs normalized to volume at days (D) 3, 8, 13, 20, and 25 post-partum, each dot represent a biological replicate, n = 5 mice/time point. Kruskal–Wallis test with Dunn’s multiple comparison: For D25 vs D3/D8/D13: ns p-value >0.9999, for D25 vs D20: ns p-value = 0.8492; Median shown. Channels and CV as labelled in image. e, Representative IF image showing normal germ-free animal liver lobule metabolic zonation depicting glutamine synthetase (GS), acetolactate synthase (ALS), argininosuccinate synthase 1 (ASS1) and Cytochrome P450 2E (Cyp2e1) expression, for comparison: see SPF zonation gradients in Fig. 1b. f–k, Representative IF images showing F4/80+ KC distribution within mouse liver (insets showing F4/80+ staining only) and total number of KCs normalized to volume in GF and SPF animals (each dot present a biological replicate) (f, g), n = 5 mice/condition; Two-Tailed Mann–Whitney test, ns p-value = 0.5159, Median shown; GF animals after cohousing with SPF animals (each dot present a biological replicate) (h–i), n = 6 mice/condition, Kruskal–Wallis test with Dunn’s multiple comparison. For SPF vs D4/D14: ns p-value >0.9999, for SPF vs D2 ns p-value = 0.2650, for SPF vs D7 ns p-value = 0.5255; Median shown. SPF animals after antibiotic (ABX) treatment, (each dot presents a biological replicate) (j–k), n = 6 mice/condition, Kruskal–Wallis test with Dunn’s multiple comparison. For SPF vs 3-wks: ns p-value = 0.8725, for SPF vs 6-wks: ns p-value = 0.2936; Median shown. l, Representative IF image showing F4/80+ KC distribution of GF animals orally gavaged with LPS (two weeks post treatment), and PBS control animals (insets showing F4/80 staining only, CV highlighted by dashed circles in IFs). m, Ratio of total KC numbers in CV to PP ROIs, each dot represents a lobule. n = 6 mice/condition, Two-tailed Mann–Whitney test, ****P < 0.0001; Median and quartiles shown. n, Total number of KCs normalized to volume in global knockouts for MyD88, TRL4, TRIF and Caspase1/11 (each dot presents a biological replicate), n = 4 Caspase 1/11−/− mice, n = 5 MyD88−/− and WT mice, n = 6 TLR4−/− and TRIF−/− animals, Kruskal–Wallis test with Dunn’s multiple comparison test. For WT vs TRIF−/−/Caspase 1/11−/−: ns p-value >0.9999, for WT vs MyD88−/−: ns p-value = 0.0776, WT vs TLR4−/−: ns p-value = 0.0524; Median shown. Channels shown labelled in figures, CV highlighted by dashed circles in IFs.
Extended Data Fig. 4 iCdh5-Cre, Alb-Cre and LysM-Cre liver reporters: loss of MyD88 signalling in hepatocytes and Kupffer cells does not alter Kupffer cell localization.
a, Representative IF image showing iCdh5-Cre expression when crossed to a Rosa26-tdTomato reporter. b, Zoomed in image: iCdh5-Cre-tdTomato is predominantly found on CD138+ LSECs, but can also be seen on a small number of KCs. c, Percentage of total tdTomato+ KCs in iCdh5-Cre-tdTomato animals, each dot represents a biological replicate, n = 3 mice. d, MFI of tdTomato on tdTomato+ F4/80+ KCs and tdTomato+ LSECs from iCdh5-Cre-tdTomato (each dot represents a cell), n = 3 mice/condition, Two-tailed Mann–Whitney test, ****P < 0.0001; Median shown. e, f, Representative IF image showing F4/80+ KC distribution of Alb-MyD88−/− (e) and LysM-MyD88−/− (f) animals with littermate controls, CV highlighted by dashed circles in IFs. g, Ratio of total KC numbers in CV to PP ROIs, (each dot represents a lobule), n = 4 mice/condition, Kruskal–Wallis test with Dunn’s multiple comparison test. For WT vs LysMCre-MyD88flfl-: ns p-value >0.9999, for WT vs AlbCre-MyD88fl/fl: ns p-value = 0.2082; Median shown. h, total KC numbers per volume; each dot represents a lobule. n = 4 mice/condition, Kruskal–Wallis test with Dunn’s multiple comparison test. For WT vs LysMCre-MyD88flfl-: ns p-value = 0.0781, for WT vs AlbCre-MyD88fl/fl: ns p-value = 0.0624; Median shown. Channels shown labelled in figures, CV highlighted by dashed circles in IFs.
Extended Data Fig. 5 Sort strategy for LSEC populations and CXCL9 translation and protein expression in iCdh5-Myd88wt/wt, iCdh5-Myd88fl/fl and Cxcl9−/− mice.
a, Representative IF image of CD117 expression around the CV on iCdh5-MyD88wt/wt, Global MyD88−/− and iCdh5-MyD88fl/fl animals, n = 2 mice. CV highlighted by dashed circles in IFs. b, Representative flow-cytometry gating strategy for bulk RNA sequencing sort: cells were sorted on forward-side scatter, singlets and live. KCs were identified by expression of F4/80+ and Tim4+. LSECs were identified by CD31+ and sorted into low, medium and high CD117 populations. c, Chemokine expression across the liver lobule as a correlate to CD117 in iCdh5-MyD88wt/wt animals. Statistical significance determined by edge-R glmLTR test, mean shown from 3 biological replicates; *p-values for: Cxcl9 = 0.009785, Ccl17 = 0.0147, Cxcl13 = 0.03433, Ccl24 = 0.01569, Ccl6 = 0.0004875. d, Representative IF image showing CXCL9-RFP expression on CD138+ LSECs in REX3 animals (Density plot of CXCL9 shown in Fig. 3a). e, Non-parametric estimate of the spatial intensity of KC, CXCL9, and CD138, (ρ(E-cadherin)) as a value of spatial covariate E-cadherin MFI, see Methods for details. n = 3 mice/condition. f, g, Log counts per million of CD117 (ns p-value = 0.9695) (f) and MyD88 (ns p-value = 1, ****P = 3.549e-23) (g) in LSECs and KCs. Statistical significance determined by edge-R glmLTR test, each dot represents a biological sample from sorted populations (n = 3 mice/condition). h, Chemokine expression across the liver lobule as a correlate to CD117 in iCdh5-MyD88fl/fl animals. In red are chemokines that are PP associated in WT animals. Statistical significance determined by edge-R glmLTR test; *p-values for: Cxcl11 = 0.01102, Ccl21a = 0.03187. i, Ratio of total KC numbers in CV to PP ROIs in global CCR2, CCR5, CCL5, CCL2, CX3CR1 knockout animals (each dot represents a lobule), n = 4 mice/condition for knockout animals, and n = 3 mice for WT controls. Kruskal–Wallis test with Dunn’s multiple comparison test, for all comparisons: ns p-value >0.9999, with the exception of WT vs CCL5−/−: ns p-value = 0.3098; Median shown. Channels shown labelled in figures, CV highlighted by dashed circles in IFs.
Extended Data Fig. 6 Expression of Kupffer cell putative markers in iCdh5-Myd88wt/wt and iCdh5-Myd88fl/fl mice.
a, Log counts per million of Cxcr3 in KCs and LSECs from MyD88wt/wt and iCdh5-MyD88fl/fl mice; biological replicates of sorted populations shown, statistical significance determined by edge-R glmLTR test; for all: ns p-value-values LSECs = 0.3191, and ns p-value-values KCs = 1. b–d, CXCR3 expression on KCs of WT, CXCR3−/− and CXCL9−/− mice as determined by flow-cytometry: Histogram of CXCR3 (b), quantification of CXCR3 MFI (c) and % of F4/80+ CXCR3+ of total F4/80+ cells (each dot represents a biological replicate) (d). n = 4 mice/group; Kruskal–Wallis test with Dunn’s multiple comparison, *P = 0.0310, **P = 0.0089; Median shown. e, Representative IF image of Tim4, Clec4f and F4/80 expression in KCs of iCdh5-MyD88wt/wt and iCdh5-MyD88fl/fl animals. CV highlighted by dashed circles in IFs. f–i, Quantification of percent of Tim4+ Clec4f+ F4/80+ cells (ns p-value = 0.5) (f) and MFI of Tim4 (ns p-value = 0.7) (g), Clec4f (ns p-value = 0.4) (h), and F4/80 (ns p-value >0.9999) (i); n = 3 mice/condition, Two-tailed Mann–Whitney test. j, Log counts per million of Nr1h3, Id3, Clec4f, Ccr2, Spic, and Timd4 in KCs from MyD88wt/wt and iCdh5-MyD88fl/fl mice; biological replicates of sorted populations shown, statistical significance determined by edge-R glmLTR test; for all: ns p-value-values = 1. k, Percent of counts associated with cell cycle from RNASeq in KCs and LSECs from MyD88wt/wt and iCdh5-MyD88fl/fl mice, biological replicates shown, Two-Way Anova with Sidak MC test, ns p-value-value for KCs = 0.4710, for LSECs = 0.1591.
Extended Data Fig. 7 RNA sequencing GAG pathway and histological ECM composition.
a, GSEA enrichment scores in relation to CD117 expression in LSECs from iCdh5-MyD88wt/wt (x-axis) and iCdh5-MyD88fl/fl (y-axis). Highlighted, pathways enriched towards the PP regions of iCdh5-MyD88wt/wt disrupted in iCdh5-MyD88fl/fl animals (Top 10 pathways shown in Fig. 3k). b, GAG pathway enrichment score in iCdh5-MyD88wt/wt and iCdh5-MyD88fl/fl. iCdh5-MyD88wt/wt show strong PP GAG enrichment, lost and/or disrupted in iCdh5-MyD88fl/fl animals. c, Representative IF image of Hyaluronic acid binding protein (HABP) detecting Hyaluronic acid (HA) in iCdh5-MyD88wt/wt and iCdh5-MyD88fl/fl. d, Representative IF image of HS in iCdh5-MyD88wt/wt and iCdh5-MyD88fl/fl. e, f, MFI of HS and HABP (respectively) in PP and CV ROIs of iCdh5-MyD88wt/wt and iCdh5-MyD88fl/fl animals, each dot represents an ROI; n = 4 iCdh5-MyD88wt/wt mice and n = 5 iCdh5-MyD88fl/fl animals, Two-Way ANOVA with Sidak multiple comparison test; Median shown. **P = 0.0011, ns p-value-value = 0.0654 (e). *P = 0.0348, ns p-value-value = 0.5972 (f). Channels shown labelled in figures.
Extended Data Fig. 8 In vivo and in vitro capture of L. monocytogenes and malaria sporozoite spatial location after liver infection.
a, Representative IF image showing L. monocytogenes-GFP captured by KCs in iCdh5-MyD88wt/wt either in PP Region (E-cadherin+) or CV Region (E-cadherin-) two hours post i.v. administration. Quantification and experimental details shown in Fig. 4a. b, c, Percent of L. monocytogenes captured in liver, spleen and blood after portal vein injection in CXCR3−/− animals (**P = 0.0023, *P = 0.0263, ns p-value = 0.7585) (b) and CXCL9−/− animals (***P = 0.0001, *P = 0.0102, ns p-value = 0.209) (c); each dot represents a biological replicate, n = 4-8 animals/group, Two-way ANOVA with Sidak multiple; Median shown. d–f, In vitro bacteria capture assay of L. monocytogenes-GFP using KCs extracted from iCdh5-MyD88wt/wt and iCdh5-MyD88fl/fl animals. Representative flow cytometry histograms of infected and uninfected KCs at 4 °C and 37 °C showing L. monocytogenes-GFP intensity (d). Quantification of flow-cytometry data showing percent of KCs with bound L. monocytogenes at 4 °C (e) and 37 °C (f); each dot represents a biological replicate, n = 4 mice/condition; Two-tailed Mann–Whitney test, ns p-value = 0.3429; Median shown. g, Representative examples of sporozoites (spz) locations one-day post liver infection in naïve animals; spz stained with anti-CSP antibody. h, i, Quantification of location of spz infection: % of total liver in WT mice that is E-cadherin+, each dot represents a biological replicate, median shown (h); % of total spz either in E-cadherin+ regions or outside E-cadherin+ regions (i); n = 250 spz imaged, data pooled from 3 different mice, each dot represents a mouse (see Methods section regarding quantification); Two-tailed Mann–Whitney (ns p-value = 0.1000).
Extended Data Fig. 9 In silico model of bacterial capture in sinusoidal network.
a, Example of the exponential distribution used to sample the KC locations along the sinusoid. The two histograms refer to two different values of the mean of the exponential distribution: λ = 25 (red- simulating a WT distribution) and λ = 500 (blue- simulating a KO distribution). b, Parameters used to constrain model simulations, obtained from analysing confocal images, mean number displayed (n = number of measurements made for each parameter, δ = standard deviation of measurements made). c, Illustrations of two example simulations in which the KC locations are sampled from an exponential distribution with mean λ = 25 (left) and λ = 500 (right). The top part of the graph corresponds to the PP region, the bottom part to the CV. White rectangles represent sinusoidal segments. Arrows schematically indicate the splitting and merging of vessels. Red rectangles symbolize KCs, while small coloured rectangles are representative of bacteria. The path of each bacterium is described by the string of rectangles of the same colour. All bacteria start their motion at the top of the graph (PP region) and only bacteria that are not successfully stopped by KCs reach the bottom (CV). In both simulations we show the path of 10 bacteria. d–f, Simulating varying KC distributions (λ) and KC bacterial binding probability in parameter space: the network’s capturing capacity is proportional to increases of KC binding probability while inversely proportional to increases of λ values. Average percent of stopped bacteria as a function of KC binding probability (KC-BP - a probabilistic value of KC capture from 0-1), and of the mean of the exponential distribution λ. Results reported in a heat map in which the colour corresponds to the average percentage of stopped bacteria, as detailed by the colour bar, rows correspond to KC-BP values and columns to different λ-values (d). Results reported as line graphs with average percentages of stopped bacteria reported and increasing values of KC binding probability (KC-BP) (e) or λ-values (f). Colour of each line corresponds to either changes of KC binding probability or λ-value, respectively.
Extended Data Fig. 10 Multiparameter, iterative staining (IBEX) of Prime and Target vaccinated animals.
a, Representative IF image of Prime and Target vaccinated animal showing expression of a diverse set of immune and non-parenchymal associated markers in relationship to E-cadherin acquired via IBEX staining (Channels shown labelled in figures). b, Ratio of total cell numbers in CV to PP ROIs, each dot represents a lobule (each dot represents a lobule); n = 3 mice/condition, Kruskal–Wallis test, Dunn’s multiple comparison test; Median shown. KC vs Desmin: ns p-value = 0.3071, KC vs CD11c: ns p-value = 0.0549, KC vs Monocytes: ns p-value = 0.0510. c, Total number of Trm (CD8+ CD44+ CXCR6+ CD69+ T-cells) per volume in iCdh5-MyD88wt/wt, iCdh5-MyD88fl/fl and CXCL9−/− animals (each dot represents a biological replicate). n = 3 mice/condition; Kruskal–Wallis test, Dunn’s multiple comparison test; Median shown, ns p-value >0.9999. d, Ratio of Trm numbers in CV to PP ROIs, each dot represents a lobule. n = 3 mice/condition; Median shown, Kruskal–Wallis test, Dunn’s multiple comparison test, **P = 0.0035, ns p-value = 0.1275.
Supplementary information
Supplementary Table 1
List of antibodies used for all immune-histochemistry and flow-cytometry experiments.
Supplementary Video 1
NKT cell patrolling behaviour in liver sinusoids. Tiled image of intravital video showing NKT cells migrating along the hepatic sinusoids of a CXCR6-GFP-/+ animal i.v. injected with E-cadhering-A594 antibody. (20X magnification, frame rate 25hertz; movie plays twice).
Supplementary Video 2
Large volumetric Ce3D microscopy of mouse liver lobule. Ce3D-processed liver sample of LysM-tdTomato CXCR6-GFP dual reporter mice, E-cadherin (cyan) demarking the PP-regions and Collagen IV (gray) the liver sinusoids.
Supplementary Video 3
Visualization of multiparameter, iterative staining (IBEX) of Prime-target vaccinated animals. Single colour panels from iterative staining performed by IBEX, each panel is shown one at a time.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gola, A., Dorrington, M.G., Speranza, E. et al. Commensal-driven immune zonation of the liver promotes host defence. Nature 589, 131–136 (2021). https://doi.org/10.1038/s41586-020-2977-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2977-2
This article is cited by
-
A hepatic network of dendritic cells mediates CD4 T cell help outside lymphoid organs
Nature Communications (2024)
-
A spatiotemporal atlas of mouse liver homeostasis and regeneration
Nature Genetics (2024)
-
Locally sourced: site-specific immune barriers to metastasis
Nature Reviews Immunology (2023)
-
IBEX: a user-friendly and open-source solution for high-plex immunostaining
Nature Reviews Immunology (2023)
-
Gut–liver axis: barriers and functional circuits
Nature Reviews Gastroenterology & Hepatology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.