Abstract
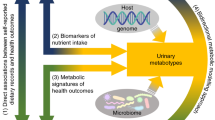
The serum metabolome contains a plethora of biomarkers and causative agents of various diseases, some of which are endogenously produced and some that have been taken up from the environment1. The origins of specific compounds are known, including metabolites that are highly heritable2,3, or those that are influenced by the gut microbiome4, by lifestyle choices such as smoking5, or by diet6. However, the key determinants of most metabolites are still poorly understood. Here we measured the levels of 1,251 metabolites in serum samples from a unique and deeply phenotyped healthy human cohort of 491 individuals. We applied machine-learning algorithms to predict metabolite levels in held-out individuals on the basis of host genetics, gut microbiome, clinical parameters, diet, lifestyle and anthropometric measurements, and obtained statistically significant predictions for more than 76% of the profiled metabolites. Diet and microbiome had the strongest predictive power, and each explained hundreds of metabolites—in some cases, explaining more than 50% of the observed variance. We further validated microbiome-related predictions by showing a high replication rate in two geographically independent cohorts7,8 that were not available to us when we trained the algorithms. We used feature attribution analysis9 to reveal specific dietary and bacterial interactions. We further demonstrate that some of these interactions might be causal, as some metabolites that we predicted to be positively associated with bread were found to increase after a randomized clinical trial of bread intervention. Overall, our results reveal potential determinants of more than 800 metabolites, paving the way towards a mechanistic understanding of alterations in metabolites under different conditions and to designing interventions for manipulating the levels of circulating metabolites.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The raw metagenomic sequencing data are available from the European Nucleotide Archive under accession numbers PRJEB11532, PRJEB17643, and for TwinsUK PRJEB32731. The raw metabolomics data and the phenotypic data are available from the European Genome–phenome Archive (EGA; https://ega-archive.org/) with accession number EGAS00001004512. Known links between genetic loci and serum metabolites were taken from the GWAS Catalog46 (https://www.ebi.ac.uk/gwas/) and the GWAS server2,22 (http://metabolomics.helmholtz-muenchen.de/gwas/).
Code availability
Source code for analysis is available at https://github.com/noambar/SerumMetabolomePredictions.
References
Psychogios, N. et al. The human serum metabolome. PLoS ONE 6, e16957 (2011).
Shin, S.-Y. et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 46, 543–550 (2014).
Long, T. et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nat. Genet. 49, 568–578 (2017).
Wikoff, W. R. et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl Acad. Sci. USA 106, 3698–3703 (2009).
Xu, T. et al. Effects of smoking and smoking cessation on human serum metabolite profile: results from the KORA cohort study. BMC Med. 11, 60 (2013).
Playdon, M. C. et al. Comparing metabolite profiles of habitual diet in serum and urine. Am. J. Clin. Nutr. 104, 776–789 (2016).
Moayyeri, A., Hammond, C. J., Hart, D. J. & Spector, T. D. The UK Adult Twin Registry (TwinsUK Resource). Twin Res. Hum. Genet. 16, 144–149 (2013).
Koivula, R. W. et al. Discovery of biomarkers for glycaemic deterioration before and after the onset of type 2 diabetes: rationale and design of the epidemiological studies within the IMI DIRECT Consortium. Diabetologia 57, 1132–1142 (2014).
Lundberg, S. M., Erion, G. G. & Lee, S.-I. Consistent individualized feature attribution for tree ensembles. Preprint at https://arxiv.org/abs/1802.03888v3 (2018).
Zeevi, D. et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015).
Yousri, N. A. et al. Long term conservation of human metabolic phenotypes and link to heritability. Metabolomics 10, 1005–1017 (2014).
Ke, G. et al. LightGBM: a highly efficient gradient boosting decision tree. In Advances in Neural Information Processing Systems 30 (eds Guyon, I. et al.) (Neural Information Processing Systems Foundation, 2017).
Cirulli, E. T. et al. Profound perturbation of the metabolome in obesity is associated with health risk. Cell Metab. 29, 488–500.e2 (2019).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Azam, S., Hadi, N., Khan, N. U. & Hadi, S. M. Antioxidant and prooxidant properties of caffeine, theobromine and xanthine. Med. Sci. Monit. 9, BR325–BR330 (2003).
Tsutsumi, Y. et al. Renal disposition of a furan dicarboxylic acid and other uremic toxins in the rat. J. Pharmacol. Exp. Ther. 303, 880–887 (2002).
Prentice, K. J. et al. CMPF, a metabolite formed upon prescription omega-3-acid ethyl ester supplementation, prevents and reverses steatosis. EBioMedicine 27, 200–213 (2018).
Nemet, I. et al. A cardiovascular disease-linked gut microbial metabolite acts via adrenergic receptors. Cell 180, 862–877.e22 (2020).
Hung, S.-C., Kuo, K.-L., Wu, C.-C. & Tarng, D.-C. Indoxyl sulfate: A novel cardiovascular risk factor in chronic kidney disease. J. Am. Heart Assoc. https://doi.org/10.1161/JAHA.116.005022 (2017).
Evenepoel, P., Meijers, B. K. I., Bammens, B. R. M. & Verbeke, K. Uremic toxins originating from colonic microbial metabolism. Kidney Int. 76, S12–S19 (2009).
Yousri, N. A. et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nat. Commun. 9, 333 (2018).
Suhre, K. et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature 477, 54–60 (2011).
Gieger, C. et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 4, e1000282 (2008).
Kettunen, J. et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat. Genet. 44, 269–276 (2012).
Capo-chichi, C. D. et al. Riboflavin and riboflavin-derived cofactors in adolescent girls with anorexia nervosa. Am. J. Clin. Nutr. 69, 672–678 (1999).
Darst, B. F., Lu, Q., Johnson, S. C. & Engelman, C. D. Integrated analysis of genomics, longitudinal metabolomics, and Alzheimer’s risk factors among 1,111 cohort participants. Genet. Epidemiol. 43, 657–674 (2019).
Sookoian, S., Castaño, G., Gianotti, T. F., Gemma, C. & Pirola, C. J. Polymorphisms of MRP2 (ABCC2) are associated with susceptibility to nonalcoholic fatty liver disease. J. Nutr. Biochem. 20, 765–770 (2009).
Hamaguchi, M. et al. Aging is a risk factor of nonalcoholic fatty liver disease in premenopausal women. World J. Gastroenterol. 18, 237–243 (2012).
Korem, T. et al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 25, 1243–1253.e5 (2017).
Olthof, M. R., van Vliet, T., Boelsma, E. & Verhoef, P. Low dose betaine supplementation leads to immediate and long term lowering of plasma homocysteine in healthy men and women. J. Nutr. 133, 4135–4138 (2003).
Craig, S. A. S. Betaine in human nutrition. Am. J. Clin. Nutr. 80, 539–549 (2004).
Liu, J. et al. Integration of epidemiologic, pharmacologic, genetic and gut microbiome data in a drug-metabolite atlas. Nat. Med. 26, 110–117 (2020).
Rothschild, D. et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 555, 210–215 (2018).
Segata, N. et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat. Methods 9, 811–814 (2012).
Li, J. et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32, 834–841 (2014).
Zeevi, D. et al. Structural variation in the gut microbiome associates with host health. Nature 568, 43–48 (2019).
Evans, A. M. et al. High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Metabolomics 4, 1000132 (2014).
Zierer, J. et al. The fecal metabolome as a functional readout of the gut microbiome. Nat. Genet. 50, 790–795 (2018).
Marco-Sola, S., Sammeth, M., Guigó, R. & Ribeca, P. The GEM mapper: fast, accurate and versatile alignment by filtration. Nat. Methods 9, 1185–1188 (2012).
Pasolli, E. et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell 176, 649–662.e20 (2019).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Korem, T. et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 349, 1101–1106 (2015).
Efron, B. & Tibshirani, R. J. An Introduction to the Bootstrap (Chapman and Hall, 1994).
Fisher, R. A. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika 10, 507 (1915).
Wald, A. Sequential tests of statistical hypotheses. Ann. Math. Stat. 16, 117–186 (1945).
Buniello, A. et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019).
Lundberg, S. & Lee, S.-I. A unified approach to interpreting model predictions. Preprint at https://arxiv.org/abs/1705.07874v2 (2017).
Manor, O. & Borenstein, E. Systematic characterization and analysis of the taxonomic drivers of functional shifts in the human microbiome. Cell Host Microbe 21, 254–267 (2017).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Yang, J., Lee, S. H., Goddard, M. E. & Visscher, P. M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Schweiger, R. et al. RL-SKAT: an exact and efficient score test for heritability and set tests. Genetics 207, 1275–1283 (2017).
Acknowledgements
We thank past and present members of the Segal group for discussions. N.B. received a PhD scholarship for Data Science by the Israeli Council for Higher Education (CHE) via the Weizmann Data Science Research Center and is supported by a research grant from Madame Olga Klein Astrachan. T.K. is a CIFAR Azrieli Global Scholar in the Humans & the Microbiome Program. E.S. is supported by the Crown Human Genome Center, by D. L. Schwarz, J. N. Halpern and L. Steinberg, and by grants funded by the European Research Council and the Israel Science Foundation. The work leading to this publication has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no.115317 (DIRECT), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and in-kind contribution from EFPIA companies. We thank A. Dutta for introducing us to the DIRECT consortium dataset.
Author information
Authors and Affiliations
Consortia
Contributions
N.B. and T.K. conceived the project, designed and conducted all analyses, interpreted the results and wrote the manuscript, and are listed in arbitrary order. O.W. and D.Z. designed statistical analyses. D.R. and S.L. conducted microbiome analysis. N.K. coordinated and designed data collection. M.L.-P. and A.W. developed protocols, performed microbiome sequencing and processed serum samples. A.W. designed the project and oversaw sample collection and processing. C.I.L.R., C.M., A.V., M.F. and T.D.S. performed the replication analysis on the TwinsUK cohort. J.A., P.W.F. and O.P. performed the replication analysis on the IMI DIRECT cohort. E.S. conceived and directed the project and analyses, designed the analyses, interpreted the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Katherine Pollard, Eric Topol and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Accurate and reproducible untargeted serum metabolomics.
a, Breakdown of the 1,251 measured metabolites by type. b, Number of samples (y axis) in which each metabolite (x axis) was identified, sorted by prevalence. c, d, Mass-spectrometry measurements (y axis) versus standardized lab tests results (x axis; Methods) for creatinine (c; Pearson’s R = 0.87, P < 10−20) and cholesterol (d; R = 0.79, P < 10−20). e, Spearman correlations (y axis: centre, median; box, IQR; whiskers, 1.5 × IQR) between standardized metabolomic profiles (Methods) of different individuals (n = 475; median Spearman’s ρ 0.05, s.d. 0.12) stratified by sex, and between standardized metabolomic profiles of the same participant (n = 20; median Spearman ρ 0.68, s.d. 0.06) taken one week apart. C&V, cofactors and vitamins; a.u., arbitrary units.
Extended Data Fig. 2 Biological sub pathway prediction of unidentified molecules.
Figure panels refer to the results of a leave-one-out cross validation prediction model of sub pathways of metabolites based on their normalized levels, raw mean, percentage of missing values, and SHAP values (Methods). Results shown are for a model trained using only sub pathways that include over 10 molecules in our data (28 sub pathways, 572 named metabolites). a, The overall accuracy of the sub pathway classifier (y axis) when a success is considered as having the true label in one of the top k predictions (x axis). b, The log loss of the classifier (y axis) computed over the resulting soft max (raw probabilities; blue) and a dichotomous matrix in which for every metabolite we only keep the top predicted sub pathway as 1 and zero-out all other predictions (red). c, The overall accuracy of the model (left y axis; blue) and the corresponding fraction of metabolites (right y axis; red) when considering only metabolites for which the classifier predicted a maximal probability above some threshold (x axis). d, A confusion matrix showing the predicted sub pathways (x axis), determined as the label with the highest probability per metabolite, versus the true annotated sub pathways (y axis). Each cell in the matrix counts the number of metabolites of a certain true sub pathway (y axis) which were assigned with some predicted sub pathway (x axis) by our model. The rightmost column is the sum of every row and represents the number of metabolites annotated for every sub pathway. The matrix is ordered by the higher order biological pathway (super pathway). Cell colours are log scaled. e, Classification results summarizing the f1-score, precision and recall per sub pathway. Rows correspond to the sub pathway annotation in d. f, For every sub pathway (y axis) shown are the fraction of metabolites truly annotated as such (black), predicted as such by the classifier (blue; out of the named molecules in the support of the model), and the fraction of unidentified molecules predicted as such (out of all unidentified molecules). M., metabolism; Xeno., xenobiotics; Ptds, peptides; AAs, amino acids.
Extended Data Fig. 3 Comparative analysis of linear versus nonlinear models and in-sample versus out-of-sample predictions.
a, Metabolite prediction R2 of GBDT versus Lasso regression models using diet data. Shown are only metabolites for which at least one model achieved significant predictions with R2 greater than 0.05. b, Histogram of the differences between the R2 of GBDT compared to Lasso regression using the diet data. c, The levels of the metabolite hydroxy-CMPF* (y axis: centre, median; box, IQR; whiskers, 1.5 × IQR) versus the monthly consumption of cooked, baked or grilled fish as reported in a food frequency questionnaire. The comparison of Spearman’s and Pearson’s correlation coefficients suggests that the relationship between the metabolite and the numerical values of the question are monotonic yet nonlinear, which explains why GBDT performs better in predicting the levels of hydroxy-CMPF* from diet data. The x axis is not to scale. d, e, Same as a, b for microbiome. f, Estimations of gut microbiome explainability (b2) of metabolite levels obtained via applying a linear mixed model on the bacterial species composition as previously described (y axis) versus the explained variance (R2) of metabolites from out-of-sample prediction models based on the same gut microbiome data. Shown are only metabolites with significant b2 estimates (5% FDR). g, Histogram of the differences between the b2 estimates and the R2 of out-of-sample prediction using the gut microbiome data.
Extended Data Fig. 4 Comparison of explained variance of metabolites for every pair of feature groups.
Dot plots of the explained variance of the metabolite groups (y axis) from models based on every pair of feature groups (x axis). Panels on the diagonal show the marginal distribution of explained variance of metabolite groups for a certain feature group.
Extended Data Fig. 5 Comparative analysis of different feature groups.
a, Spearman correlations computed between the EV of metabolites for every pair of feature groups. b, The proportion of variance explained by each of the first 400 principal components (left y axis; black) and their cumulative EV (right y axis; blue). c, R2 multiplied by the sign of the Pearson correlation coefficient (x axis) between metabolite levels and BMI in our study, versus the mean R2 multiplied by the sign of the Pearson correlation coefficient (y axis) of BMI associated metabolites recently reported by a different group13. Shown are 36 (out of 49) BMI associated metabolites that were also measured in this cohort. P value for the Pearson correlation, P = 7 × 10−11. Line and shaded colouring represent the fitting of a linear model and the 95% confidence interval. d, The EV of every metabolite from prediction models based on the gut microbiome (x axis) versus diet (y axis). Dashed red line is y = x. e, Same for prediction models based on both gut microbiome and diet (x axis) compared to using only diet (y axis). f, Same for prediction models based on diet and permuted gut microbiome (x axis) compared to using only diet (y axis). MB, microbiome.
Extended Data Fig. 6 Networks of interactions between phenotypes explain diverse metabolites.
Interactions between features from different feature groups predictive of similar metabolites are presented in a graphical layout, in which nodes are either metabolites or features, and edges are the directional mean absolute SHAP values (Methods) computed from models trained only on features from the respective feature group. Circular nodes, metabolites; predictive feature nodes, squares; both coloured by relevant categories. Shown are only edges with a mean absolute SHAP value greater than 0.12. a, Network of associations for the following feature groups: macronutrients, diet, microbiome, lifestyle, drugs and seasonal effects. b, A large group of metabolites for which predictions are mainly driven by the reported consumption of coffee and the relative abundance of a bacteria from the Clostridiales order. c, Metabolites explained by seasonal fruit consumption. d, Selected examples of interactions between metabolites and features in predictive models.
Extended Data Fig. 7 Specific dietary features and bacterial taxa underlie the accurate prediction of circulating metabolites.
a–f, Predicted (y axis) versus measured (x axis) levels (arbitrary units) of X-16124 (a; Pearson’s R = 0.77, P < 10−20), phenylacetylglutamine (b; R = 0.63, P < 10−20), p-cresol-glucuronide (c; R = 0.64, P < 10−20), caffeine (d; R = 0.68, P < 10−20), hydroxy-CMPF (e; R = 0.72, P < 10−20) and stachydrine (f; R = 0.5, P < 10−20). Predictions of a–c are based only on microbiome data, and coloured by the relative abundance of the bacterial taxa having the highest mean absolute SHAP value for each metabolite. Predictions of d–f are based only on diet data, and coloured by the reported consumption of the dietary item having the highest mean absolute SHAP value for each metabolite. P values for prediction were estimated via bootstrapping. g, Heat map showing the directional mean absolute SHAP values (Methods) of various features (x axis) computed from fivefold cross validation models that predict metabolite levels (y axis) using two separate models, one based on diet and another on gut microbiome data. Positive (negative) SHAP values indicate that higher (lower) feature values lead, on average, to higher predicted values. Shown are the top 150 predicted metabolites using diet and gut microbiome, and the top 40 features by maximum mean absolute SHAP value across all metabolites.
Extended Data Fig. 8 Distribution of phyla and a taxa from the Eggerthellaceae family.
a, Stacked bar plots per sample (x axis) showing the relative abundance of bacterial phyla (y axis). Samples are sorted by the relative abundance of the most abundant phylum, Firmicutes. Bacteroidetes is the second most abundant phylum in our cohort. Relative abundance of a phylum is computed as the sum over relative abundances of all bacterial features belonging to that phylum. b, The levels of the unidentified compound X-16124 in individuals for which the bacterial taxa from the Eggerthellaceae family was detectable in stool versus individuals for which it was not (P < 10−20, two-sided Mann–Whitney U-test).
Extended Data Fig. 9 The unidentified molecule X-21441 associates with rs8187710 independent of age.
a, A table showing the coefficients, standard errors and P values resulted from a multiple linear regression model with levels of the unidentified molecule X-21441 as the dependent variable, the allele dosage of rs8187710 (0–2) and age (years) as the independent variables: yX-21441 = constant + β1 × rs8187710 + β2 × Age. b, The levels of X-21441 (y axis; centre, median; box, IQR; whiskers, 1.5 × IQR) versus the genotype of the participants (x axis). The number of participants with each genotype is indicated below the tick labels. The explained variance of X-21441 by rs8187710 as estimated using plink (Methods) is indicated on the upper right corner of the panel. c, The levels of X-21441 versus the age of the participants (x axis) coloured by genotype of participants. Line and shaded colouring represent the fitting of a linear model and the 95% confidence interval. SE, standard error.
Supplementary information
Supplementary Information
This file contains Supplementary Notes 1-4.
Supplementary Tables
This file contains Supplementary Tables 1-15.
Rights and permissions
About this article
Cite this article
Bar, N., Korem, T., Weissbrod, O. et al. A reference map of potential determinants for the human serum metabolome. Nature 588, 135–140 (2020). https://doi.org/10.1038/s41586-020-2896-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2896-2
This article is cited by
-
Gut microbiome-metabolome interactions predict host condition
Microbiome (2024)
-
Metabolomic profiling of cancer-related fatigue involved in cachexia and chemotherapy
Scientific Reports (2024)
-
Variant of the lactase LCT gene explains association between milk intake and incident type 2 diabetes
Nature Metabolism (2024)
-
Multi-omic integration of microbiome data for identifying disease-associated modules
Nature Communications (2024)
-
Identification of blood metabolites associated with risk of Alzheimer’s disease by integrating genomics and metabolomics data
Molecular Psychiatry (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.