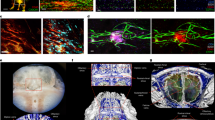
Abstract
The central nervous system has historically been viewed as an immune-privileged site, but recent data have shown that the meninges—the membranes that surround the brain and spinal cord—contain a diverse population of immune cells1. So far, studies have focused on macrophages and T cells, but have not included a detailed analysis of meningeal humoral immunity. Here we show that, during homeostasis, the mouse and human meninges contain IgA-secreting plasma cells. These cells are positioned adjacent to dural venous sinuses: regions of slow blood flow with fenestrations that can potentially permit blood-borne pathogens to access the brain2. Peri-sinus IgA plasma cells increased with age and following a breach of the intestinal barrier. Conversely, they were scarce in germ-free mice, but their presence was restored by gut re-colonization. B cell receptor sequencing confirmed that meningeal IgA+ cells originated in the intestine. Specific depletion of meningeal plasma cells or IgA deficiency resulted in reduced fungal entrapment in the peri-sinus region and increased spread into the brain following intravenous challenge, showing that meningeal IgA is essential for defending the central nervous system at this vulnerable venous barrier surface.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Korin, B. et al. High-dimensional, single-cell characterization of the brain’s immune compartment. Nat. Neurosci. 20, 1300–1309 (2017).
Schuchardt, F. et al. In vivo analysis of physiological 3D blood flow of cerebral veins. Eur. Radiol. 25, 2371–2380 (2015).
Louveau, A. et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015).
Rua, R. & McGavern, D. B. Advances in meningeal immunity. Trends Mol. Med. 24, 542–559 (2018).
Herz, J., Filiano, A. J., Smith, A., Yogev, N. & Kipnis, J. Myeloid cells in the central nervous system. Immunity 46, 943–956 (2017).
Polfliet, M. M. et al. Meningeal and perivascular macrophages of the central nervous system play a protective role during bacterial meningitis. J. Immunol. 167, 4644–4650 (2001).
Gitlin, A. D., Shulman, Z. & Nussenzweig, M. C. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 509, 637–640 (2014).
Moser, K., Tokoyoda, K., Radbruch, A., MacLennan, I. & Manz, R. A. Stromal niches, plasma cell differentiation and survival. Curr. Opin. Immunol. 18, 265–270 (2006).
Mora, J. R. & von Andrian, U. H. Differentiation and homing of IgA-secreting cells. Mucosal Immunol. 1, 96–109 (2008).
Henriksson, A., Kam-Hansen, S. & Link, H. IgM, IgA and IgG producing cells in cerebrospinal fluid and peripheral blood in multiple sclerosis. Clin. Exp. Immunol. 62, 176–184 (1985).
Roberg, M., Forsberg, P., Tegnell, A. & Ekerfeldt, K. Intrathecal production of specific IgA antibodies in CNS infections. J. Neurol. 242, 390–397 (1995).
Doss, S. et al. High prevalence of NMDA receptor IgA/IgM antibodies in different dementia types. Ann. Clin. Transl. Neurol. 1, 822–832 (2014).
Westman, G. et al. Clinical significance of IgM and IgA class anti-NMDAR antibodies in herpes simplex encephalitis. J. Clin. Virol. 103, 75–80 (2018).
Rojas, O. L. et al. Recirculating intestinal IgA-producing cells regulate neuroinflammation via IL-10. Cell 176, 610–624.e618 (2019).
Biajoux, V. et al. Efficient plasma cell differentiation and trafficking require Cxcr4 desensitization. Cell Rep. 17, 193–205 (2016).
Senda, S., Cheng, E. & Kawanishi, H. Aging-associated changes in murine intestinal immunoglobulin A and M secretions. Scand. J. Immunol. 27, 157–164 (1988).
Hapfelmeier, S. et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science 328, 1705–1709 (2010).
Kamada, N. et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012).
Knutton, S. et al. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17, 2166–2176 (1998).
Schnupf, P. et al. Growth and host interaction of mouse segmented filamentous bacteria in vitro. Nature 520, 99–103 (2015).
Casteleyn, C., Rekecki, A., Van der Aa, A., Simoens, P. & Van den Broeck, W. Surface area assessment of the murine intestinal tract as a prerequisite for oral dose translation from mouse to man. Lab. Anim. 44, 176–183 (2010).
Moor, K. et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502 (2017).
Wills, S. et al. HIV-1-specific IgA monoclonal antibodies from an HIV-1 vaccinee mediate galactosylceramide blocking and phagocytosis. J. Virol. 92, e01552-17 (2018).
Bai, X. D., Liu, X. H. & Tong, Q. Y. Intestinal colonization with Candida albicans and mucosal immunity. World J. Gastroenterol. 10, 2124–2126 (2004).
Bidgood, S. R., Tam, J. C., McEwan, W. A., Mallery, D. L. & James, L. C. Translocalized IgA mediates neutralization and stimulates innate immunity inside infected cells. Proc. Natl Acad. Sci. USA 111, 13463–13468 (2014).
Chakrabarti, A. Epidemiology of central nervous system mycoses. Neurol. India 55, 191–197 (2007).
Limon, J. J., Skalski, J. H. & Underhill, D. M. Commensal fungi in health and disease. Cell Host Microbe 22, 156–165 (2017).
Moran, E. et al. Proteasome inhibitors as immunosuppressants: biological rationale and clinical experience. Semin. Hematol. 49, 270–276 (2012).
Mastorakos, P. & McGavern, D. The anatomy and immunology of vasculature in the central nervous system. Sci. Immunol. 4, eaav0492 (2019).
Fagarasan, S. Evolution, development, mechanism and function of IgA in the gut. Curr. Opin. Immunol. 20, 170–177 (2008).
Rua, R. et al. Infection drives meningeal engraftment by inflammatory monocytes that impairs CNS immunity. Nat. Immunol. 20, 407–419 (2019).
St Leger, A. J. et al. An ocular commensal protects against corneal infection by driving an interleukin-17 response from mucosal γδ T cells. Immunity 47, 148–158.e145 (2017).
Manglani, M. & McGavern, D. B. Intravital imaging of neuroimmune interactions through a thinned skull. Curr. Protoc. Immunol. 120, 24.2.1–24.2.12 (2018).
Naik, S. et al. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature 520, 104–108 (2015).
Watson, S. J. et al. Viral population analysis and minority-variant detection using short read next-generation sequencing. Phil. Trans. R. Soc. Lond. B 368, 20120205 (2013).
Lefranc, M. P. et al. IMGT, the international ImMunoGeneTics information system, http://imgt.cines.fr. Nucleic Acids Res. 37, D1006–D1012 (2009).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Brochet, X., Lefranc, M. P. & Giudicelli, V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36, W503–W508 (2008).
Gupta, N. T. et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015).
Bashford, G. R., Burnfield, J. M. & Perez, L. C. Physical activity discrimination improvement using accelerometers and wireless sensor network localization - biomed 2013. Biomed. Sci. Instrum. 49, 243–250 (2013).
Shugay, M. et al. VDJtools: unifying post-analysis of t cell receptor repertoires. PLOS Comput. Biol. 11, e1004503 (2015).
Forster, S. C. et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat. Biotechnol. 37, 186–192 (2019).
Acknowledgements
The Clatworthy laboratory is based in the University of Cambridge Molecular Immunity Unit in the MRC Laboratory of Molecular Biology and is grateful for the use of the core facilities. J.F. and M.R.C. are supported by the National Institute of Health Research (NIHR) Cambridge Biomedical Research Centre and the NIHR Blood and Transplant Research Unit, and M.R.C. by a Medical Research Council New Investigator Research Grant (MR/N024907/1), a Chan-Zuckerburg Initiative Human Cell Atlas Technology Development Grant, a Versus Arthritis Cure Challenge Research Grant (21777), and an NIHR Research Professorship (RP-2017-08-ST2-002). D.B.M., D.S.R., M.L.N.-D. and Z.F. are supported by the Intramural Research Program of NINDS. Y.B. and N.B. are supported by the Intramural Research Program of NIAID. We thank R. Barker for discussions; A. Thomas (NCI) and G. Nair (NINDS) for human dura mater; the NIAID Microbiome Program gnotobiotic animal facility staff (D. Trageser-Cesler and C. Acevedo); and S. Krishnamurthy for his work contributing to the colonization of germ-free mice with commensal bacteria.
Author information
Authors and Affiliations
Contributions
M.R.C. conceived the project. Z.F., D.B.M. and M.R.C. designed experiments, interpreted results and wrote the manuscript with input from all authors. Y.B. and T.D.L. designed and interpreted intestinal colonization experiments. G.F., A.F. and Z.F. conducted and analysed results from DSS experiments. J.F. and A.F. analysed bulk RNA sequencing data. J.F. and O.S. generated the BCR sequencing data, which were analysed by Z.K.T. with advice from R.B.-R. M.L.N.-D. and Z.F. conducted intravital two-photon microscopy experiments. N.K. analysed gut microbiome sequencing of DA and DB mouse lines. S.C., N.B., K.H. and Z.F. conducted intestinal colonization experiments. T.T., A.H., D.S.R. and Z.F. acquired and analysed human dura mater samples. K.S. provided advice on experimental protocol. Z.F. conducted and analysed results from all additional experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Richard Flavell and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Anatomical localization of dural B cells and IgA+ cells.
a, Representative flow cytometric plot of meningeal CD19+B220+ cells from a naive B6 mouse with a histogram depicting CD79b expression on this population of cells relative to CD11b+ myeloid cells. The flow cytometric plot depicts the proportion of naive (IgD+IgM+) and class-switched (CS, IgD-IgM-) B cells of total CD19+B220+ CD79b+ cells, which is quantified in the bar graph (mean \(\pm \) s.d., n = 5 mice). b, Representative image of a meningeal whole-mount from a Prdm1EYFP/+ mouse stained with IgA (red) and CD138 (blue) showing co-localization with Blimp1-EYFP+ cells (green) (scale bar, 50 μm). Inset represents a higher magnification of the highlighted box (scale bar, 30 μm); the dotted white line depicts the lumen of the transverse sinus (TS). c, Image of a meningeal whole-mount from a naive B6 mouse immunolabelled for IgA (red) and B220 (green) displaying the localization of tissue-resident B cells and IgA+ cells in the dura mater (scale bar, 1,000 μm). The arrows depict the TS and superior sagittal sinus (SSS). d, Normalized counts from RNA-seq of meninges from SPF and GF mice for the indicated genes. Normalized counts calculated using DESeq (n = 3 mice per group as a box plot). e, Representative image of the peri-SSS region from a naive B6 mouse stained for CXCR4 (red), B220 (green) and IgA (blue) (scale bar, 50 μm). f, Representative image of IgA (red) and CD138 (green) immunolabelling of whole-mount human dura mater in the lobe area with DAPI (blue, 4′,6-diamidino-2-phenylindole) in blue (scale bar, 30 μm).
Extended Data Fig. 2 Bacterial diversity in the gut of DA-GF and DB-GF mouse lines.
a, b, Comparison of microbial richness using Chao1 (a) and diversity using Inverse Simpson indexes (b) between two groups (DA-GF: blue, n = 6; DB-GF: green, n = 6) is shown. Violin plots are density plots where the width indicates frequency and the box plot shows median, 95% CI and IQR (two-sided Wilcoxon signed-rank test). c, Log twofold change in the relative abundance of taxa between two groups (DA-GF: blue, n = 6; DB-GF: green, n = 6). Only taxa that are significantly different between two groups are shown (Kruskal–Wallis test).
Extended Data Fig. 3 Clonal relatedness of IgA+ cells in the gut and meninges of SPF, DA GF and DB GF mice, and meningeal Ig heavy chain gene expression in DSS colitis mice.
a, Hypergeometric test for enrichment (over-representation) of the shared clones between small intestines and meninges. An overlap occurring by random chance would be <13% or <154 clones (dotted black line). The observed overlap in clones between meninges and small intestine = 237/1,103 clones (red dotted line) and is non-random (P = 4.2 × 10−21). b, Percentage of lineage trees with most ancestral sequences beginning from meninges (M) or small intestines (SI) in SPF mice inferred after BCR lineage reconstruction. Statistical analysis was performed using an unpaired two-tailed Student’s t-test where P < 0.05 was considered statistically significant. c, Diversity estimation of CDR3 repertoire represented as a calculated Chao1 estimate after subsampling with 1,000 random iterations (left panel). Wilcoxon rank-sum test was used to determine statistical significance (P < 0.05 considered statistically significant). Right, relative proportions (frequency of reads) of unique CDR3 amino acid sequences present in each SPF sample is shown, and the top ten most frequent sequences are coloured in decreasing shades of grey. Clones after top 10 are not coloured (white). d, (Left panels) BCR network of meninges and small intestines from a representative SPF mouse. Colours indicate isotype of BCR sequences. (Right panels) BCR network of IGA clones only from the same mouse. Shared clones between meninges and small intestines are highlighted with the same colours in both networks. Grey nodes and edges indicate BCR sequences/clones not shared between the two tissues. e, Relative proportions (frequency of reads) of unique CDR3aa sequences present in each DA-GF and DB-GF sample is shown, and the top ten most frequent CDR3aa sequences are coloured in decreasing shades of grey. Clones after top 10 are not coloured (white). f, BCR network of IGA clones from meninges and small intestines in SPF (left), DA-GF (middle), and DB-GF (right) mice. BCR clones shared between meninges and small intestines are highlighted with the same colours in both networks. Grey nodes and edges indicate BCR sequences/clones not shared between the two tissues. g, Normalized counts for the indicated Ig heavy chain genes from bulk RNA-seq of meninges from either DSS-colitic mice or control mice (n = 6 mice per group). Note that, with exception to IGHA (Extended Data Fig. 4d), heavy chain transcript expression did not significantly differ between DSS and control mice.
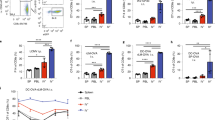
Extended Data Fig. 4 Accumulation and proliferation of meningeal IgA+ cells after gut-epithelial barrier breach.
a, Representative confocal images of IgA (red) and B220 (green) immunolabelling and Phalloidin staining (grey) of the SSS from a mouse receiving normal drinking water and from a mouse receiving 2% DSS in drinking water for 7 days followed by normal water for 2 weeks to induce acute colitis. (scale bar, 200 μm); the inset is a higher magnification of the highlighted box depicting a cluster of B cells and IgA+ cells associating adjacent to Lyve-1+ lymphatic vessels (yellow) (scale bar, 40 μm). b, Quantification of peri-sinus IgA+ cells from control vs 2% DSS mice (mean \(\pm \) s.d., n = 5 mice per group; **P = 0.0079, two-tailed Mann–Whitney test). c, Quantification of B220+ cells along the SSS in control vs 2% DSS mice (mean \(\pm \) s.d., n = 4 mice in control group and 5 mice in DSS group; *P = 0.0159, two-tailed Mann–Whitney test). d, Normalized counts of meningeal Igha expression determined by RNA-seq in the denoted groups (n = 6 mice per group). e, A representative image of the TS region of whole-mount tissue from a mouse receiving 2% DSS in drinking water for 7 days followed by normal water for 2 weeks to induce acute colitis. Tissue was stained for B220 (green), Ki-67 (red) and DAPI (blue) (scale bar, 50 μm). The inset to the right depicts a cluster of proliferating B cells (scale bar, 20 μm).
Extended Data Fig. 5 B cell expansion in meninges following systemic candidiasis.
a, Representative flow cytometric plot of CD19+B220+CD79b+ meningeal B cells after intravenous administration of C. albicans, with a bar graph to the right depicting an increase in the total number of naive (IgD+IgM+) and class-switched (CS, IgD−IgM−) B cells 3 days after infection (mean \(\pm \) s.d., n = 4 mice per group Total CD19+B220+CD79b+ B cells **P = 0.0074, Total IgD+IgM+ B cells *P = 0.0103, Total IgD−IgM− B cells **P = 0.0040, unpaired two-tailed Student’s t-test). b, Representative image of peri-sinus immune cell clusters from whole-mount tissue stained with IgA (white), B220 (red) and CD45 (green) from a mouse 2 days post systemic candida infection showing GFP-C. albicans in purple (scale bar, 50 μm). The inset represents a higher magnification image of the highlighted region displaying IgA+ cells and diffuse IgA signal in regions with pathogen sequestration (scale bar, 20 μm); yellow arrowhead depicts an extracellular GFP-C. albicans cell, whereas the turquoise arrowhead shows GFP-C. albicans signal within a CD45+ leukocyte. c, Representative image of the superior sagittal sinus (SSS) region from the indicated groups, stained for B220 (green) and Ki-67 (red), showing a peri-SSS cluster of Ki-67+B220+ cells 3 days after infection (scale bar, 50 μm). The inset to the right represents a higher magnification of the highlighted box. d, Representative image of the SSS region in a B6 mouse one week after systemic candida infection immunolabelled for IgA (blue), B220 (green) and AID (white). The yellow arrowheads depict AID-expressing IgA+ cells and the red arrowhead depicts AID-expressing B220+ cells (scale bar, 40 μm).
Extended Data Fig. 6 Distribution of C. albicans in IgA-deficient mice.
a, Quantification of peri-sinus IgA+ cells in wild type (WT) vs IgA-deficient animals by confocal imaging of IgA-stained whole-mount tissue (n = 3 mice per group). b, Representative images of the SSS immunolabelled for IgA (green) from a wild type (WT) vs IgA-deficient mouse 2 days after i.v. administration of GFP-C. albicans shown in purple (scale bar, 150 μm). Quantification of GFP-C. albicans+ puncta along the SSS normalized by area in each group is shown in the bar graph (mean \(\pm \) s.d., n = 3 mice per group, **P = 0.0065, unpaired two-tailed Student’s t-test). c, Representative images captured in the hippocampus of a WT vs IgA-deficient mouse stained for Iba1 (red) 2 days after i.v. administration of GFP-C. albicans (shown in green; white arrowheads) (scale bar, 50 μm). Quantification of hippocampal GFP-C. albicans+ puncta normalized by area is shown in the bar graph. Each data point represents the average of four serial hippocampal images from a single animal (mean \(\pm \) s.d., n = 4 WT mice and 6 IgA−/− mice, *P = 0.0428, unpaired two-tailed Student’s t-test).
Extended Data Fig. 7 Local effects of bortezomib-mediated meningeal plasma cell depletion.
a, Schematic paradigm of sub-scalp administration of either vehicle control hydrogel (Gel) or bortezomib-containing hydrogel (Bz) for meningeal plasma cell depletion. b, Representative histo-cytometry dot plots from meningeal whole-mounts immunolabelled for IgA (red), B220 (green) and CD3 (blue) from either a mouse injected with Gel or Bz. c, Representative confocal images of IgA (red) and Iba1 (green) immunolabelling in whole-mount meninges of the SSS region, depicting the presence of peri-sinus IgA+ cells and macrophages in a mouse administered either Gel or Bz (scale bar, 100 μm). d, Quantification of peri-sinus IgA+ cells in mice injected with either Gel or Bz, as assessed by immunohistochemical analysis of whole-mount tissue (mean \(\pm \) s.d., n = 4 mice per group; **P = 0.0014, unpaired two-tailed Student’s t-test). e, Quantification of peri-sinus B220+ cells (B cells), CD3+ cells (T cells) and Iba1+ cells (macrophages) from immunolabelling of meningeal whole-mounts in mice administered either Gel or Bz. Total cell counts were normalized by area (mean \(\pm \) s.d., n = 4 mice per group; B220+ cells *P = 0.0473, CD3+ cells P = 0.2897, Iba1+ cells P = 0.3716, unpaired two-tailed Student’s t-test; ns, not significant). f, Gating strategy for flow cytometric analysis of meningeal immune cell subsets following Bz treatment. g, Bar graph depicting the total cell counts of the indicated immune cell populations in Gel- or Bz-treated animals (mean \(\pm \) s.d., n = 4 mice per group; no statistically significant changes were noted in each group).
Extended Data Fig. 8 Effects of bortezomib-mediated meningeal plasma cell depletion on bone marrow and spleen compartments.
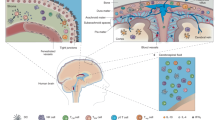
a, Gating strategy for flow cytometric analysis of spleen and bone marrow plasma cells in the protocol depicted in Extended Data Fig. 5b. b, Representative flow cytometric plots showing CD138+ plasma cells in the spleen from either Gel- or Bz-treated mice. c, Quantification of absolute numbers of splenic plasma cells by flow cytometry in either Gel- or Bz-treated mice (mean \(\pm \) s.d., n = 4 mice per group; P = 0.4693, unpaired two-tailed Student’s t-test; ns, not significant). d, Representative FACS plots of bone marrow plasma cells and IgA+ plasma cells in either Gel or Bz treated mice. e, Flow cytometric quantification of absolute numbers of total bone marrow plasma cells and IgA+ plasma cells in Gel- or Bz-treated mice (mean \(\pm \) s.d., n = 4 mice per group; Total CD138+B220− cells P = 0.1369, Total CD138+B220−IgA+ cells P = 0.1556, unpaired two-tailed Student’s t-test; ns, not significant). f, Illustration highlighting the role of gut-educated IgA+ plasma cells along the dural sinuses in entrapping blood-borne microbes and protecting the brain from infection.
Supplementary information
Supplementary Table
This file contains Supplementary Table 1: BCR Primers.
Video 1
Meningeal plasma cell dynamics under steady-state conditions. Two-photon microscopy was used to capture a time-lapse movie of meningeal EYFP-Blimp1+ cells (green) by imaging a naive Prdm1EYFP/+ mouse under a thinned skull. EYFP-Blimp1+ cells can be seen closely associating with peri-sinus vasculature (red), which was made apparent by an intravenous injection of Evans blue. The field of view is just below the anterior SSS (scale bar, 10 μm).
Video 2
Peri-sinus B cell and plasma cell clusters during systemic candidiasis. After induction of Cre recombinase, a CD19CreER/+ Stopfl/flTdTomato x Prdm1EYFP/+ mouse was infected by intravenous administration of 5 × 104 viable C. albicans cells and imaged 5 days later by two-photon microscopy through a thinned skull. Highlighted by the arrowheads, clusters of CD19+ B cells (red) with associating CD19+Blimp1+ plasma cells (yellow) can be seen along the SSS, which is outlined by the dotted line (scale bar, 40 μm).
Rights and permissions
About this article
Cite this article
Fitzpatrick, Z., Frazer, G., Ferro, A. et al. Gut-educated IgA plasma cells defend the meningeal venous sinuses. Nature 587, 472–476 (2020). https://doi.org/10.1038/s41586-020-2886-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2886-4
This article is cited by
-
Role of meningeal immunity in brain function and protection against pathogens
Journal of Inflammation (2024)
-
Unraveling the diversity and functions of tissue-resident plasma cells
Nature Immunology (2024)
-
What Jane Jacobs has taught me about neuroimmunology
Nature Immunology (2024)
-
Neuroimmunology of Cardiovascular Disease
Current Hypertension Reports (2024)
-
Slc43a2+ T cell metastasis from spleen to brain in RGNNV infected teleost
Science China Life Sciences (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.