Abstract
Muscle regeneration is sustained by infiltrating macrophages and the consequent activation of satellite cells1,2,3,4. Macrophages and satellite cells communicate in different ways1,2,3,4,5, but their metabolic interplay has not been investigated. Here we show, in a mouse model, that muscle injuries and ageing are characterized by intra-tissue restrictions of glutamine. Low levels of glutamine endow macrophages with the metabolic ability to secrete glutamine via enhanced glutamine synthetase (GS) activity, at the expense of glutamine oxidation mediated by glutamate dehydrogenase 1 (GLUD1). Glud1-knockout macrophages display constitutively high GS activity, which prevents glutamine shortages. The uptake of macrophage-derived glutamine by satellite cells through the glutamine transporter SLC1A5 activates mTOR and promotes the proliferation and differentiation of satellite cells. Consequently, macrophage-specific deletion or pharmacological inhibition of GLUD1 improves muscle regeneration and functional recovery in response to acute injury, ischaemia or ageing. Conversely, SLC1A5 blockade in satellite cells or GS inactivation in macrophages negatively affects satellite cell functions and muscle regeneration. These results highlight the metabolic crosstalk between satellite cells and macrophages, in which macrophage-derived glutamine sustains the functions of satellite cells. Thus, the targeting of GLUD1 may offer therapeutic opportunities for the regeneration of injured or aged muscles.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) data repository, with accession number GSE123825. Other data that support the findings of this study are available from the corresponding author (M.M.) upon reasonable request. Source data are provided with this paper.
References
Bentzinger, C. F., Wang, Y. X., Dumont, N. A. & Rudnicki, M. A. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 14, 1062–1072 (2013).
Costamagna, D., Berardi, E., Ceccarelli, G. & Sampaolesi, M. Adult stem cells and skeletal muscle regeneration. Curr. Gene Ther. 15, 348–363 (2015).
Saclier, M., Cuvellier, S., Magnan, M., Mounier, R. & Chazaud, B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 280, 4118–4130 (2013).
Saclier, M. et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells 31, 384–396 (2013).
Tidball, J. G. Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 17, 165–178 (2017).
Perdiguero, E. et al. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J. Cell Biol. 195, 307–322 (2011).
Latroche, C. et al. Coupling between myogenesis and angiogenesis during skeletal muscle regeneration is stimulated by restorative macrophages. Stem Cell Reports 9, 2018–2033 (2017).
Summan, M. et al. Macrophages and skeletal muscle regeneration: a clodronate-containing liposome depletion study. Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R1488–R1495 (2006).
Rennie, M. J. et al. Skeletal muscle glutamine transport, intramuscular glutamine concentration, and muscle–protein turnover. Metabolism 38 (Suppl 1), 47–51 (1989).
Biolo, G., Fleming, R. Y., Maggi, S. P. & Wolfe, R. R. Transmembrane transport and intracellular kinetics of amino acids in human skeletal muscle. Am. J. Physiol. 268, E75–E84 (1995).
Nurjhan, N. et al. Glutamine: a major gluconeogenic precursor and vehicle for interorgan carbon transport in man. J. Clin. Invest. 95, 272–277 (1995).
Palmieri, E. M. et al. Pharmacologic or genetic targeting of glutamine synthetase skews macrophages toward an M1-like phenotype and inhibits tumor metastasis. Cell Rep. 20, 1654–1666 (2017).
St Pierre, B. A. & Tidball, J. G. Differential response of macrophage subpopulations to soleus muscle reloading after rat hindlimb suspension. J. Appl. Physiol. 77, 290–297 (1994).
Guardiola, O. et al. Induction of acute skeletal muscle regeneration by cardiotoxin injection. J. Vis. Exp. 119, e54515 (2017).
Takeda, Y. et al. Macrophage skewing by Phd2 haplodeficiency prevents ischaemia by inducing arteriogenesis. Nature 479, 122–126 (2011).
von Maltzahn, J., Jones, A. E., Parks, R. J. & Rudnicki, M. A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl Acad. Sci. USA 110, 16474–16479 (2013).
Zammit, P. S. Function of the myogenic regulatory factors Myf5, MyoD, myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis. Semin. Cell Dev. Biol. 72, 19–32 (2017).
Wenes, M. et al. Macrophage metabolism controls tumor blood vessel morphogenesis and metastasis. Cell Metab. 24, 701–715 (2016).
Yang, C. et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56, 414–424 (2014).
Rodgers, J. T. et al. mTORC1 controls the adaptive transition of quiescent stem cells from G0 to GAlert. Nature 510, 393–396 (2014).
Zhang, P. et al. mTOR is necessary for proper satellite cell activity and skeletal muscle regeneration. Biochem. Biophys. Res. Commun. 463, 102–108 (2015).
Jewell, J. L. et al. Differential regulation of mTORC1 by leucine and glutamine. Science 347, 194–198 (2015).
Rayagiri, S. S. et al. Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat. Commun. 9, 1075 (2018).
Sousa-Victor, P. et al. Geriatric muscle stem cells switch reversible quiescence into senescence. Nature 506, 316–321 (2014).
Bernet, J. D. et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat. Med. 20, 265–271 (2014).
Jin, L. et al. Glutamate dehydrogenase 1 signals through antioxidant glutathione peroxidase 1 to regulate redox homeostasis and tumor growth. Cancer Cell 27, 257–270 (2015).
Liu, P. S. et al. α-Ketoglutarate orchestrates macrophage activation through metabolic and epigenetic reprogramming. Nat. Immunol. 18, 985–994 (2017).
Obara, H., Matsubara, K. & Kitagawa, Y. Acute limb ischemia. Ann. Vasc. Dis. 11, 443–448 (2018).
Sayer, A. A. et al. New horizons in the pathogenesis, diagnosis and management of sarcopenia. Age Ageing 42, 145–150 (2013).
Vinciguerra, M., Musaro, A. & Rosenthal, N. Regulation of muscle atrophy in aging and disease. Adv. Exp. Med. Biol. 694, 211–233 (2010).
Carobbio, S. et al. Deletion of glutamate dehydrogenase in β-cells abolishes part of the insulin secretory response not required for glucose homeostasis. J. Biol. Chem. 284, 921–929 (2009).
He, Y. et al. Glutamine synthetase deficiency in murine astrocytes results in neonatal death. Glia 58, 741–754 (2010).
Mingote, S. et al. Genetic pharmacotherapy as an early CNS drug development strategy: testing glutaminase inhibition for schizophrenia treatment in adult mice. Front. Syst. Neurosci. 9, 165 (2016).
Guardiola, O. et al. Cripto regulates skeletal muscle regeneration and modulates satellite cell determination by antagonizing myostatin. Proc. Natl Acad. Sci. USA 109, E3231–E3240 (2012).
LaFleur, M. W. et al. A CRISPR–Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 10, 1668 (2019).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Pasut, A., Jones, A. E. & Rudnicki, M. A. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp. 73, e50074 (2013).
Casazza, A. et al. Impeding macrophage entry into hypoxic tumor areas by Sema3A/Nrp1 signaling blockade inhibits angiogenesis and restores antitumor immunity. Cancer Cell 24, 695–709 (2013).
Acknowledgements
M.M. was supported by an ERC Consolidator grant (ImmunoFit), FWO-SBO (ZL3C3602), Horizon 2020 (research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 766214). We thank V. van Hoef for bioinformatic analyses; S.-M. Fendt, C. Frezza, A. Musarò, G. Cossu and J.-C. Marine for advice; and S. Trusso Cafarello and S. Willox for technical support. P.C. and M.M. received long-term structural Methusalem funding by the Flemish Government; P.C. is supported by an ERC PoC (ERC-713758) and Advanced grant (EU-ERC743074). M. Shang received a grant from the China Scholarship Council (CSC); E.B. received a grant from the FWO (1525315N).
Author information
Authors and Affiliations
Contributions
M. Shang performed experimental design, all experiments, data acquisition and interpretation, and wrote the manuscript. F.C. and R.A. performed in vitro assays and histology. J.S. performed all the ligations and histological staining. F.V. performed angiogenic and in vitro assays. M.Y.R. provided AAV vectors. G.E. performed Seahorse measurements. S.C. and P.M. generated GLUD1 conditional knockout mice and provided critical suggestions. K.D.B. provided the transgenic mice expressing CreERT under the Pax7 promoter, and provided critical edits to the text. M. Sandri provided critical edits to the text. P.-C.H. helped in the experiments with GLS-knockout macrophages and provided GlsΔMo mice. B.G. and P.C. supported with metabolic assays and critical suggestions in manuscript writing. M.D.M. designed and supervised all the in vitro and in vivo gene editing approaches, and provided critical edits to the text. E.B. and M.M. performed the experimental design and data analysis, conducted scientific direction and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Terry Partridge and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Infiltrating GLUD1-deficient macrophages improve muscle repair.
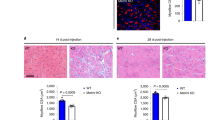
a, Western blot for GLUD1 in BMDMs from control and Glud1ΔMo mice. Vinculin was used as loading control. Representative image of three independent blots. b, c, RT–qPCR of Glud1 in F4/80+ macrophages (b), and Glud1 in Ly6G+ neutrophils (c), sorted from tibialis anterior muscles 1 day post-CTX (n = 4). d, Monocyte-derived macrophages (F4/80+GFP−) and tissue-resident macrophages (F4/80+GFP+) in tibialis anterior muscles 1 day post-CTX. Injured mice were CD68.eGFP transgenic mice reconstituted with wild-type (WT→CD68.eGFP) (n = 3) or Glud1ΔMo bone marrow cells (Glud1 KO→CD68.eGFP) (n = 4). e, Necrotic area on H&E-stained sections from tibialis anterior muscles 6 days post-CTX. Injured mice were CD68.eGFP transgenic mice reconstituted with wild-type (WT→CD68.eGFP) or Glud1ΔMo bone marrow cells (KO→CD68.eGFP) (n = 6). Baseline (B.): WT→CD68.eGFP (n = 3); KO→CD68.eGFP (n = 4). f, RT–qPCR of Glud1 in F4/80+ macrophages, sorted from spleens upon tamoxifen-induced macrophage-specific Glud1 deletion in Glud1L/L;Csf1rcreERT mice (designated L/L in the figure); tamoxifen-injected littermates (Glud1L/L and negative for Csf1rcreERT; designated WT in the figure) were used as controls (n = 5).g–i, Quantification of necrosis (g), apoptosis (h) and regenerating fibres (i), from tibialis anterior muscles 6 days post-CTX in tamoxifen-injected Glud1L/L;Csf1rcreERT mice and littermate controls (Glud1L/L and negative for Csf1rcreERT) (n = 6). j–l, Quantification of proliferating (Ki67-expressing) satellite cells in tibialis anterior muscles (j) 1 day post-CTX injury (control, n = 4; Glud1ΔMo n = 5) with representative images (k), or in crural muscles (l) 3 days post-ligation (n = 5). The yellow arrows indicate PAX7+Ki67− cells, and the white arrows indicate PAX7+Ki67+ cells. m, n, Western blot for PAX7 in tibialis anterior muscles lysates (m) from control or Glud1ΔMo mice 1 day post-CTX (n = 4), and densitometric quantification (n). Vinculin was used as loading control. Numbers represent fold change versus vinculin. o–u, FACS quantification of total CD45+ leukocytes (o), F4/80+ macrophages (p), Ly6G+ neutrophils (q), TCRβ+ total T cells (r), CD4+ T cells (s), CD8+ cytotoxic T cells (t) and CD45R+ B cells (u), in tibialis anterior muscles at baseline or 1 day post-CTX (n = 3). v, Laser Doppler analysis 1, 3, 6, 9 and 13 days post-ligation (control n = 5 for all the time points; Glud1ΔMo, day 0, 1, 3 and 6, n = 4; day 9 and 13, n = 3). Toe perfusion of nonligated control was defined as 100%. w, Quantification of vessel density in crural muscles 14 days post-ligation (control, n = 5; Glud1ΔMo, n = 3). A representative (a–m, o–w) or a pool (n) of at least two independent experiments is shown. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 50 μm. Graphs show mean ± s.e.m.
Extended Data Fig. 2 GLUD1 loss in macrophages does not alter either their recruitment or M1, M2 or wound-healing gene-expression patterns.
a, Crystal-violet-stained BMDMs, migrating towards CCL21, CCL2 or PBS (Uns) in glutamine (Q)-enriched or glutamine-reduced medium (n = 3). b, c, Quantification (b) and representative images (c) of F4/80 staining in ear sections with acetone (vehicle) or upon TPA-induced cutaneous rash, 3 days and 8 days after applying TPA (vehicle, n = 4; TPA day 3, n = 6 control and 5 Glud1ΔMo; TPA day 8, n = 4). d, Heat map analysis of M1 and M2 macrophage gene expression in CD45+F4/80+ macrophages sorted from tibialis anterior muscles at baseline and 1 day post-CTX (n = 4). e, Heat map analysis of wound-healing gene expression in CD45+F4/80+ macrophages sorted from tibialis anterior muscles at baseline and 1 day post-CTX (n = 4). In a–c, experiments show representative values of two independent experiments; d, e show values from one single experiment. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 50 μm. Graphs show mean ± s.e.m.
Extended Data Fig. 3 GLUD1 loss in macrophages does not alter either M1 and M2 polarization or their related functions.
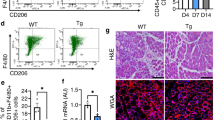
a–d, RT–qPCR of Cxcl9 (a), Tnfa (b), Arg1 (c) and Il10 (d) in BMDMs isolated from control and Glud1ΔMo mice (n = 3). e–h, FACS analysis of different M1 (e, f) or M2 (g, h) polarization states in CD45+CD11b+F4/80+ macrophages isolated from tibialis anterior muscles at baseline (n = 5) or 1 day post-CTX (n = 6). i, Quantification of macrophage phagocytosis. BMDMs were treated with LPS or PBS (unstimulated) before the assay (n = 3). j, k, Quantification (j), and representative images (k), of total endothelial sprout length of spheroid containing HUVECs and wild-type or Glud1ΔMo BMDMs. BMDMs were treated with IL-4 before the assay; unstimulated BMDMs were used as control (unstimulated n = 7; IL-4 n = 8). l, m, CD206+F4/80+ area in tibialis anterior muscles 1 day (n = 5) and 6 days (n = 8) post-CTX (l), or in crural muscles 3 days (control n = 6; Glud1ΔMo n = 5), 7 days (control n = 7; Glud1ΔMo n = 5) and 14 days (control n = 6; Glud1ΔMo n = 5) post-ligation (m). All experiments show representative values of at least two independent experiments. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 50 μm. Graphs show mean ± s.e.m.
Extended Data Fig. 4 GLUD1 loss in macrophages enhances GS-mediated glutamine release.
a, Quantification (by gas chromatography–mass spectrometry) of intracellular 2-OG content in BMDMs cultured in glutamine-enriched or glutamine-reduced medium (n = 3). b, c, Liquid chromatography–mass spectrometry measurement of total cellular energy charge ([ATP + 1/2ADP]/[ATP + ADP + AMP]) (b) and ATP content (c) in BMDMs (n = 3). d, Oxygen consumption rate (OCR) in BMDMs (n = 5). e, f, Quantification of intracellular (e) and extracellular (f) glutamine content in BMDMs cultured in glutamine-enriched or glutamine-reduced medium (n = 3). g, [U-14C]glutamine uptake in BMDMs cultured in glutamine-enriched (n = 4) or glutamine-reduced (wild type n = 4; Glud1ΔMo n = 3) medium. h, Evaluation of [U-13C]glutamine-derived carbon incorporation into glutamate in BMDMs (n = 3). i, j, Evaluation of [U-13C]glucose-derived carbon incorporation levels into 2-OG (i) and glutamate (j) in BMDMs (n = 3). k, l, Quantification of intracellular (k) and extracellular (l) glutamine content in BMDMs upon silencing of BCAT1 or BCAT2 (n = 3). m, n, Quantification of intracellular (m) and extracellular (n) glutamine content in BMDMs upon silencing of GOT1 or GOT2 (n = 3). o, Quantification of satellite cells on tibialis anterior muscles 1 day post-CTX injury, stained for pHH3 and PAX7. Control and Glud1ΔMo mice were treated two times per day with the BCAT1 inhibitor gabapentin, or vehicle as control (n = 6). p, Fold change in glutamate-to-leucine ratio in the interstitial fluid of tibialis anterior muscles 1 day post-CTX, relative to PBS-injected control muscle (PBS n = 6; CTX n = 9). q, Fold change in glutamate-to-leucine ratio in the interstitial fluid of crural muscles 3 days post-ligation, relative to control baseline muscle (baseline n = 7, 8 control, Glud1ΔMo, respectively; ligated n = 11, 12 control, Glud1ΔMo, respectively). r, Evaluation of the conversion of glutamate to 2-OG by analysing [U-13C]glutamine (glutamine-enriched condition) or [U-13C]glutamate (glutamine-reduced condition) incorporation into 2-OG in wild-type BMDMs (n = 3). s, Evaluation of the conversion of 2-OG to glutamate by analysing 15NH4+ incorporation into glutamate in wild-type BMDMs (n = 3). t, Evaluation of GS activity by analysing 15NH4+ incorporation into glutamine in BMDMs (n = 3). u, v, Evaluation of the conversion of GLUD1 activity (u), and GS activity (v), in muscle-infiltrating macrophages, sorted 1 day post-CTX. One unit for the conversion of glutamate to 2-OG is the amount of enzyme that will generate 1 μmole of NADH per minute at pH 7.6 at 37 °C. One unit of GS activity is defined as the enzyme producing 1 nmole of γ-glutamyl hydroxamic acid per minute (control n = 4; Glud1ΔMo n = 3). The control condition in u, v is the same one displayed in Fig. 2p at day 1. All experiments (except for o) show representative values of at least two independent experiments, o shows values from one single experiment. Unpaired two-tailed t-test; NS, not significant (P > 0.05); a.u., arbitrary unit. Graphs show mean ± s.e.m.
Extended Data Fig. 5 Harnessing glutamine uptake in vitro.
a, b, Quantification (a) and representative images (b) of myotube diameter in C2C12 cells cultured in BMDM-conditioned medium (n = 3 except for glutamine-reduced C2C12, for which n = 2). c, RT–qPCR of Slc1a5 knockdown efficiency in C2C12 cells. Cells were transduced with a lentivirus co-expressing Cas9 and a gRNA targeting the Slc1a5 locus (SLC1A5-KD) (n = 5) or a nontargeting control gRNA (n = 4). d, [U-14C]glutamine uptake in SLC1A5-deficient C2C12 cells (SLC1A5 KD) generated by co-expressing Cas9 along with a gRNA targeting the Slc1a5 locus. Parental cells (control) and cells transduced with a nontargeting control gRNA were used as negative controls. C2C12 cells treated with SLC1A5 inhibitor GPNA were used as a positive control (n = 3). e, f, Quantification (e) and representative images (f) of myotube diameter in control or SLC1A5-KD C2C12 cells cocultured with BMDMs under glutamine deprivation (n = 3 except control C2C12 n = 2). g, RT–qPCR analysis of the proliferation marker Pcna in control or SLC1A5-KD C2C12 cells, or control C2C12 treated with the mTOR inhibitor Torin2, cultured for 18 h in BMDM-conditioned glutamine-reduced growth medium, in which the only glutamine present comes from wild-type or GLUD1-knockout BMDMs. A nontargeting control gRNA was used as control (n = 3). h, RT–qPCR analysis of the differentiation marker Myog in control or SLC1A5-KD C2C12 cells, or control C2C12 treated with the mTOR inhibitor Torin2, cultured for 72 h in BMDM-conditioned glutamine-reduced differentiation medium, in which the only glutamine present comes from wild-type or GLUD1-knockout BMDMs. A nontargeting control gRNA was used as control (n = 3). i, Representative images of an immunofluorescence for PAX7 on a pure satellite cell population, freshly isolated from hindlimb muscles of wild-type mice. j, RT–qPCR for Slc1a5 in satellite cells, transduced with the same lentivirus as above. The graph shows values of three biological repetitions per condition. k, l, Quantification (k) and representative images (l) of EdU by immunofluorescence in control or SLC1A5-KD satellite cells. A nontargeting control gRNA was used as a control (control gRNA n = 5; SLC1A5-KD n = 6). m–o, Quantification (m, n) and representative images (o) of fusion index and myotube size in control or SLC1A5-KD satellite cells after 5 days of culture in differentiation medium. A nontargeting control gRNA was used as a control. The graph shows values of three biological repetitions per condition. All experiments show representative values of at least two independent experiments. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 50 μm (b, f, l), 100 μm (o). Graphs show mean ± s.e.m.
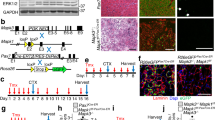
Extended Data Fig. 6 Selective and inducible knockdown of Slc1a5 in satellite cells.
a, Schematic of the AAV8 expression vector for in vivo targeting of satellite cells. U6, Pol III promoter driving the expression of the gRNA targeting the Slc1a5 locus or a nontargeting control gRNA. Because the mice used in this experiment are LSL-Cas9/Pax7creERT mice, Cas9 is exclusively activated in PAX7+ cells upon tamoxifen administration and genome editing of the Slc1a5 locus will occur selectively in satellite cells. b, Schematic of an AAV8-based CRISPR–Cas9-mediated in vivo genome editing. c, d, Representative images (c) and quantification (d) for PAX7 and Cas9 staining on uninjured muscles before and after tamoxifen administration (n = 4). e, f, RT–qPCR for Slc1a5 in freshly isolated satellite cells (n = 4) (e) and all other mononuclear cells (nonsatellite cells) (n = 3) (f) upon in vivo genome editing of the Slc1a5 locus (SLC1A5-KD) specifically in satellite cells. Nontargeting control gRNA was used as a control. g, h, Quantification (g) and representative images (h) of SLC1A5 and PAX7 staining on freshly isolated satellite cells, upon in vivo genome editing of the Slc1a5 locus (SLC1A5-KD) specifically in satellite cells (n = 3). All experiments show representative values of at least two independent experiments. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 50 μm (c), 20 μm (h). Graphs show mean ± s.e.m.
Extended Data Fig. 7 Slc1a5 knockdown in satellite cells impairs the recovery of the muscle from CTX-induced damage.
a–d, Quantification of TUNEL+ cells (a), F4/80+ area (c) and representative images (b, d), in tibialis anterior muscle 6 days post-CTX obtained from LSL-Cas9/Pax7creERT mice treated with an AAV8 vector encoding control gRNA or Slc1a5 gRNA (SLC1A5-KD) (n = 4). e, f, Quantification (e) and representative images (f) of EdU+ myonuclei in tibialis anterior muscle 6 days post-CTX, upon in vivo genome editing of the Slc1a5 locus (SLC1A5-KD) specifically in satellite cells. EdU was given by intraperitoneal injection at 24 h, 48 h and 72 h after CTX injection (n = 6). In a–d, representative values of two independent experiments are shown; e, f show values of one experiment. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 20 μm. Graphs show mean ± s.e.m.
Extended Data Fig. 8 Macrophage-specific genetic deletion or pharmacological inhibition of GLUD1 alters the basal inflammation and weight of muscle tissue only in aged mice.
a, Representative images of F4/80+ area in crural muscles of young and aged mice. b–i, Quantification and representative images of F4/80+ area in brain (b, c), liver (d, e), lung (f, g), and skin (h, i) of aged mice (n = 5 except in b for Glud1ΔMo n = 4). j–n, Body weight (j) and mass-to-body weight ratio of kidney (k), liver (l), spleen (m) and fat tissues (n) of aged mice upon R162 treatment (control n = 5; Glud1ΔMo n = 6). In a–i, representative values of at least two independent experiments are shown; j–n show values of one experiment. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 50 μm (a, i); 20 μm (c, e, g). Graphs show mean ± s.e.m.
Extended Data Fig. 9 GLS loss in macrophages is not advantageous for muscle repair.
a, b, [U-14C]glutamine uptake (a) and glutamine oxidation (b) in wild-type or GLS-knockout BMDMs cultured with glutamine-enriched or glutamine-reduced medium (n = 3). c, d, 2-OG-to-succinate ratio in wild-type or GLS-knockout BMDMs (c) and 2-OG-to-succinate ratio in wild-type or GLUD1-knockout BMDMs (d). BMDMs were treated with 50 ng/ml LPS or PBS (unstimulated) before the assay (n = 3). e, Evaluation of GS activity by analysing the percentage of the 15NH4+-derived ammonia incorporation levels into glutamine in BMDMs isolated from control and GlsΔMo mice (n = 3). f, Fold change in glutamine-to-glutamate ratio in the interstitial fluid of tibialis anterior muscle 1 day post-CTX, relative to PBS-injected control muscle (n = 6). g, h, Quantification of necrotic (right side of the graph) and regenerating (left side of the graph) areas on H&E-stained sections from tibialis anterior muscles 6 days post-CTX (n = 6) (g) and representative images (h). i, j, Quantification (i) and representative images (j) of TUNEL+ cells in tibialis anterior muscle 6 days post-CTX (n = 6). k–m, Representative images (k) and quantification of F4/80+ area (l), CD206−F4/80+ cells (M1) to CD206+F4/80+ cells (M2) ratio (m) in tibialis anterior muscles 6 days post-CTX (n = 6). n–q, RT–qPCR of Tnfa (n), Cxcl9 (o), Mrc1 (p) and Retnla (q) in BMDMs isolated from control and GlsΔMo mice. BMDMs were treated with LPS or PBS (unstimulated) before the assay (n = 3). r, Scheme illustrating the physiological role of GLUD1 in macrophages in response to muscle damage. During muscle disruption, ischaemia or ageing, interstitial glutamine drops—probably because of the loss in myofibres (a major glutamine source) and poor blood supply. Infiltrating macrophages respond to glutamine starvation by reducing their oxidative GLUD1 activity in favour of GS activity. Macrophage-derived glutamine is released and progressively fills the muscle interstitium, where it is taken up by satellite cells, promoting their proliferation and differentiation into new fibres (two processes that are favoured by glutamine-dependent mTOR activation). Towards the end of this regenerative process, the newly generated fibres will undertake glutamine production and inflammation will be progressively resolved. GLUD1-deficient macrophages are metabolically pre-adapted towards glutamine synthesis and release, thus preventing this glutamine drop. It follows that—in the case of muscle damage—macrophage-specific knockout of Glud1 or pharmacological GLUD1 blockade strengthens satellite cell activation, ultimately leading to therapeutic muscle regeneration. All experiments show representative values of at least two independent experiments. Unpaired two-tailed t-test; NS, not significant (P > 0.05). Scale bars, 20 μm (h), 10 μm (j, k). Graphs show mean ± s.e.m.
Supplementary information
Supplementary Figure
Supplementary Figure 1 | Uncropped western blot scans. The figure shows the original, uncropped scans of the western blot images displayed in Fig. 1v-x, Fig. 2e, f, s, Extended Data Fig. 1a,m.
Source data
Rights and permissions
About this article
Cite this article
Shang, M., Cappellesso, F., Amorim, R. et al. Macrophage-derived glutamine boosts satellite cells and muscle regeneration. Nature 587, 626–631 (2020). https://doi.org/10.1038/s41586-020-2857-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2857-9
This article is cited by
-
Advances in sarcopenia: mechanisms, therapeutic targets, and intervention strategies
Archives of Pharmacal Research (2024)
-
Functional polarization of tumor-associated macrophages dictated by metabolic reprogramming
Journal of Experimental & Clinical Cancer Research (2023)
-
Communications between macrophages and cardiomyocytes
Cell Communication and Signaling (2023)
-
The metabolomic plasma profile of patients with Duchenne muscular dystrophy: providing new evidence for its pathogenesis
Orphanet Journal of Rare Diseases (2023)
-
A novel cuproptosis-related diagnostic gene signature and differential expression validation in atherosclerosis
Molecular Biomedicine (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.