Abstract
Spinal cord injury in mammals is thought to trigger scar formation with little regeneration of axons1,2,3,4. Here we show that a crush injury to the spinal cord in neonatal mice leads to scar-free healing that permits the growth of long projecting axons through the lesion. Depletion of microglia in neonatal mice disrupts this healing process and stalls the regrowth of axons, suggesting that microglia are critical for orchestrating the injury response. Using single-cell RNA sequencing and functional analyses, we find that neonatal microglia are transiently activated and have at least two key roles in scar-free healing. First, they transiently secrete fibronectin and its binding proteins to form bridges of extracellular matrix that ligate the severed ends of the spinal cord. Second, neonatal—but not adult—microglia express several extracellular and intracellular peptidase inhibitors, as well as other molecules that are involved in resolving inflammation. We transplanted either neonatal microglia or adult microglia treated with peptidase inhibitors into spinal cord lesions of adult mice, and found that both types of microglia significantly improved healing and axon regrowth. Together, our results reveal the cellular and molecular basis of the nearly complete recovery of neonatal mice after spinal cord injury, and suggest strategies that could be used to facilitate scar-free healing in the adult mammalian nervous system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) (www.ncbi.nlm.nih.gov/geo) under the accession number GSE150871. Source data are provided with this paper.
References
O’Shea, T. M., Burda, J. E. & Sofroniew, M. V. Cell biology of spinal cord injury and repair. J. Clin. Invest. 127, 3259–3270 (2017).
Tran, A. P., Warren, P. M. & Silver, J. The biology of regeneration failure and success after spinal cord injury. Physiol. Rev. 98, 881–917 (2018).
Hilton, B. J. & Bradke, F. Can injured adult CNS axons regenerate by recapitulating development? Development 144, 3417–3429 (2017).
Stenudd, M., Sabelström, H. & Frisén, J. Role of endogenous neural stem cells in spinal cord injury and repair. JAMA Neurol. 72, 235–237 (2015).
Mokalled, M. H. et al. Injury-induced ctgfa directs glial bridging and spinal cord regeneration in zebrafish. Science 354, 630–634 (2016).
Zukor, K. A., Kent, D. T. & Odelberg, S. J. Meningeal cells and glia establish a permissive environment for axon regeneration after spinal cord injury in newts. Neural Dev. 6, 1 (2011).
Jin, Y. & Zheng, B. Multitasking: dual leucine zipper-bearing kinases in neuronal development and stress management. Annu. Rev. Cell Dev. Biol. 35, 501–521 (2019).
He, Z. & Jin, Y. Intrinsic control of axon regeneration. Neuron 90, 437–451 (2016).
Bregman, B. S. Spinal cord transplants permit the growth of serotonergic axons across the site of neonatal spinal cord transection. Brain Res. 34, 265–279 (1987).
Fry, E. J., Stolp, H. B., Lane, M. A., Dziegielewska, K. M. & Saunders, N. R. Regeneration of supraspinal axons after complete transection of the thoracic spinal cord in neonatal opossums (Monodelphis domestica). J. Comp. Neurol. 466, 422–444 (2003).
Schafer, D. P. et al. Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705 (2012).
Butovsky, O. et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 17, 131–143 (2014).
Hammond, T. R. et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271 (2019).
Popovich, P. G. & Hickey, W. F. Bone marrow chimeric rats reveal the unique distribution of resident and recruited macrophages in the contused rat spinal cord. J. Neuropathol. Exp. Neurol. 60, 676–685 (2001).
Elmore, M. R. et al. Colony-stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 82, 380–397 (2014).
Rudge, J. S. & Silver, J. Inhibition of neurite outgrowth on astroglial scars in vitro. J. Neurosci. 10, 3594–3603 (1990).
McKeon, R. J., Schreiber, R. C., Rudge, J. S. & Silver, J. Reduction of neurite outgrowth in a model of glial scarring following CNS injury is correlated with the expression of inhibitory molecules on reactive astrocytes. J. Neurosci. 11, 3398–3411 (1991).
Buettner, F. et al. Computational analysis of cell-to-cell heterogeneity in single-cell RNA-sequencing data reveals hidden subpopulations of cells. Nat. Biotechnol. 33, 155–160 (2015).
Li, Q. et al. Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10 (2019).
Mrdjen, D. et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395 (2018).
Jordão, M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363, eaat7554 (2019).
Keren-Shaul, H. et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290 (2017).
Tan, K. & Lawler, J. The interaction of thrombospondins with extracellular matrix proteins. J. Cell Commun. Signal. 3, 177–187 (2009).
Jaffe, E. A. & Mosher, D. F. Synthesis of fibronectin by cultured human endothelial cells. J. Exp. Med. 147, 1779–1791 (1978).
Zhou, X. et al. Microglia and macrophages promote corralling, wound compaction and recovery after spinal cord injury via Plexin-B2. Nat. Neurosci. 23, 337–350 (2020).
Miya, D. et al. Fetal transplants alter the development of function after spinal cord transection in newborn rats. J. Neurosci. 17, 4856–4872 (1997).
Conrad, S., Schluesener, H. J., Adibzahdeh, M. & Schwab, J. M. Spinal cord injury induction of lesional expression of profibrotic and angiogenic connective tissue growth factor confined to reactive astrocytes, invading fibroblasts and endothelial cells. J. Neurosurg. Spine 2, 319–326 (2005).
Stevens, B. & Schafer, D. P. Roles of microglia in nervous system development, plasticity, and disease. Dev. Neurobiol. 78, 559–560 (2018).
Pluvinage, J. V. et al. CD22 blockade restores homeostatic microglial phagocytosis in ageing brains. Nature 568, 187–192 (2019).
Yona, S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013).
Kisanuki, Y. Y. et al. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230, 230–242 (2001).
Postic, C. et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J. Biol. Chem. 274, 305–315 (1999).
Sakai, T. et al. Plasma fibronectin supports neuronal survival and reduces brain injury following transient focal cerebral ischemia but is not essential for skin-wound healing and hemostasis. Nat. Med. 7, 324–330 (2001).
Li, J., Chen, K., Zhu, L. & Pollard, J. W. Conditional deletion of the colony stimulating factor-1 receptor (c-fms proto-oncogene) in mice. Genesis 44, 328–335 (2006).
Saederup, N. et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One 5, e13693 (2010).
Jung, S. et al. Analysis of fractalkine receptor CX3CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106–4114 (2000).
Liu, K. et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 13, 1075–1081 (2010).
Zukor, K. et al. Short hairpin RNA against PTEN enhances regenerative growth of corticospinal tract axons after spinal cord injury. J. Neurosci. 33, 15350–15361 (2013).
Chen, B. et al. Reactivation of dormant relay pathways in injured spinal cord by KCC2 manipulations. Cell 174, 521–535 (2018).
Liu, Y. et al. A sensitized IGF1 treatment restores corticospinal axon-dependent functions. Neuron 95, 817–833 (2017).
Hanada, K. et al. Isolation and characterization of E-64, a new thiol protease inhibitor. Agric. Biol. Chem. 42, 523–528 (1978).
Vicuña, L. et al. The serine protease inhibitor serpinA3N attenuates neuropathic pain by inhibiting T cell-derived leukocyte elastase. Nat. Med. 21, 518–523 (2015).
Ou, J. et al. iPSCs from a hibernator provide a platform for studying cold adaptation and its potential medical applications. Cell 173, 851–863 (2018).
Acknowledgements
We thank J. Sanes and I. Benhar for discussion and S. Hegarty, J. Sanes, B. Stevens, F. Wang, K. H. Wang, C. Winter and C. Woolf for critically reading the manuscript. This study was supported by NIH R01 grants NS096294 and NS110850 (to Z.H.); NIH-NINDS R35NS111582 and a Ray W. Poppleton endowment (to P.G.P.); grants from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (to Z.H., R.K., V.S. and D.H.G.); the Craig H. Neilsen Foundation (to Y.L.); and a NIH training grant T32NS007473 (to A.M.).The Intellectual and Developmental Disabilities Research Center (IDDRC) and viral cores supported by the grants NIH HD018655 and P30EY012196 were used for this study.
Author information
Authors and Affiliations
Contributions
Y.L., X.H., P.G.P. and Z.H. conceived the project; Y.L., X.H., Q.W., A.M., B.C., Z.S., H.M., S.Z., J.Z. and A.J. performed the experiments and discussed the results; R.K., Y.Z., Z.Y., V.S. and D.H.G. participated in data analysis; Y.L., X.H. and Z.H. prepared the manuscript with inputs from all authors.
Corresponding author
Ethics declarations
Competing interests
Z.H. is an advisor of SpineX. The remaining authors declare no competing interests.
Additional information
Peer review information Nature thanks Elizabeth Bradbury, Jerry Silver and Binhai Zheng for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
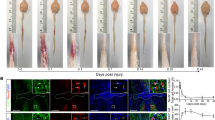
Extended Data Fig. 1 Age-dependent decline in serotonergic axon regrowth and wound healing.
a, Representative images of spinal cord sagittal sections showing 5HT-labelled axons from sham, P7 or P20 mice at two weeks after inury. Scale bar, 500 μm. b, Representative images of spinal cord sections from sham, P7 or P20 mice at two weeks after injury, stained with antibodies against collagen I, CD68, P2Y12 or CD31. Scale bar, 250 μm. c, Representative images of spinal sagittal sections at four weeks after sham control or P2 injury, with CST axons labelled with AAV-ChR2-mCherry. Red stars indicate the lesion site. Scale bar, 500 μm. d, Representative images of sagittal spinal cord sections at two weeks after injury, stained with antibodies against laminin, CSPG and GFAP. Scale bar, 250 μm. e, Representative images of sagittal spinal cord sections at two weeks after P20 injury, stained with antibodies against laminin, GFAP and 5-HT. Scale bar, 250 μm. All experiments shown were independently repeated three times with similar results.
Extended Data Fig. 2 Distinct microglia and macrophage responses after crush injury to neonatal or adult spinal cord.
a, Images of spinal cord sections stained with antibodies against CD68 and P2Y12 from mice at 3 dpi, 7 dpi or 14 dpi. Higher-magnification images showing P2Y12+ cells were co-labelled with CD68 at 3 dpi. Cells with highly ramified morphology at 7 dpi and 14 dpi can be seen around lesion sites. Scale bar, 100 μm. b, Higher-magnification images from Fig. 2a showing that CD68+ cells and fibronectin matrix form bridges between gaps at 3 dpi. Scale bar, 50 μm. c, Immunolabelling for CD68 and P2Y12 in adult mice at 3 dpi, 7 dpi or 14 dpi, showing CD68+ cells that lack P2Y12 expression. Scale bar, 200 μm. All experiments shown were independently repeated three times with similar results.
Extended Data Fig. 3 Histological assessments of bridges formed after neonatal spinal cord injury.
a, Higher-magnification images of sections of the spinal cord bridge area stained with antibodies against fibronectin, GFAP, P2Y12 and collagen I, or with DAPI (blue), at 3 dpi in P2 injury. Scale bar, 50 μm. b, Representative images of spinal sections of Cx3cr1GFP mice immunolabelled with caspase-3 showing cells around the lesion sites at 3 dpi in P2 injury. Scale bar, 200 μm. All experiments shown were independently repeated three times with similar results.
Extended Data Fig. 4 Infiltrated CCR2+ monocytes and macrophages were eliminated after neonatal but not adult spinal cord injury.
a, b, Representative images of sagittal sections of injured spinal cord of Ccr2RFP mice at 3 or 14 dpi in P2 injury (a) or adult injury (b). Sections were immunostained for CD68 and RFP (for Ccr2RFP). Scale bar, 250 μm. All experiments shown were independently repeated three times with similar results.
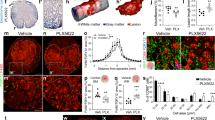
Extended Data Fig. 5 Microglia depletion impairs wound healing and axon regrowth after neonatal spinal cord injury.
a, Left, representative P2Y12-stained spinal cord images showing PLX3397-mediated depletion of microglia cells. Right, quantification of microglia depletion in spinal cord treated with PLX3397 or vehicle at 0, 7 or 14 dpi (n = 3, 5 and 5 for 0, 7 and 14 dpi, respectively). Student’s t-test (two-tailed, unpaired), ***P < 0.0001. Data are mean ± s.e.m. Scale bar, 250 μm. b, Representative images of P2Y12-stained spinal cord sections from control (Csf1rlfl/fl) and Csf1r-knockout (Cx3crcre;Csf1rfl/fl) mice showing an approximately 70% reduction of microglia throughout the spinal cord in the mutant mice (n = 5 per group). Student’s t-test (two-tailed, unpaired), ***P = 0.0004. Data are mean ± s.e.m. Scale bar, 250 μm. c, Representative images of sagittal spinal sections taken at 14 days after P2 injury and immunostained with antibodies against 5-HT, GFAP, laminin, CSPG or CD31. Scale bar, 200 μm. d, Higher-magnification images from c, showing 5-HT+ axons and GFAP+ astrocytes in the lesion site. Scale bar, 50 μm.
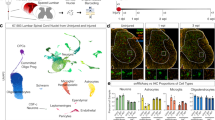
Extended Data Fig. 6 Isolation and scRNA-seq results for immune cells after neonatal spinal cord injury.
a, FACS plots showing selection of CD11b+ and CD45+ cells from dissociated cells from neonatal spinal cord. b, t-SNE plot showing 14 clusters and population annotations. c, d, Relative proportions of microglia among total cells (c) and dividing microglia among microglia cells (d). e, Left, table showing the percentage of each cluster and their signature genes. Right, heat map depicting the top 30 differentially expressed genes for each of the 14 clusters.
Extended Data Fig. 7 Feature plots showing examples of differentially expressed genes in different clusters.
Gene expression of P2ry12, Tmem119, Siglech, Fn1, Igf1, Spp1, Ms4a7, Thbs1, Anxa1, Cstb, Serpinb6a and Stfa1 in different microglia clusters.
Extended Data Fig. 8 Comparison of changes in gene expression in proliferative-region-associated microglia, disease-associated microglia and MG3.
Dot plots of gene expression correlation between proliferative-region-associated microglia (PAM) and disease-associated microglia (DAM) (normalized to homeostatic microglia) and MG3 (normalized to MG0), showing different sets of upregulated and downregulated genes (n = 13,755 and 14,423 genes for PAM and DAM, respectively).
Extended Data Fig. 9 Network analysis and further characterization of MG3 differentially expressed genes.
a, Diagram depicting correlated gene modules that underlie cluster identities of MG3 microglia. b, c, Expression of genes associated with endopeptidase inhibitor activity (b) and the ECM (c), in adult microglia at 0, 3 and 5 dpi, using bulk RNA-seq (n = 3 per group). One-way ANOVA followed by post-hoc Bonferroni correction. *P = 0.03 (Lgals3), 0.04 (Lgals1), 0.01 (Ecm1), 0.01(Pf4); **P = 0.003 (Fn1), 0.0013 (Pf4). Data are mean ± s.e.m.
Extended Data Fig. 10 Microglia isolation and transplantation.
a, Representative images (left) and quantification (right) of isolated microglia (P2Y12+) from neonatal or adult Cx3cr1GFP mice (3 h after isolation, n = 400 cells examined over three independent experiments). Data are mean ± s.e.m. Scale bar, 50 μm. b, Representative images of spinal cord sections showing the activation of transplanted microglia in the adult lesion at two days after grafting. Scale bar, 250 μm. All experiments shown in b were independently repeated three times with similar results. c, Representative images of spinal cord sections showing the GFAP and CSPG staining in the adult lesion at 14 days after grafting. Quantification results are shown in Fig. 5d. Scale bar, 250 μm.
Supplementary information
Rights and permissions
About this article
Cite this article
Li, Y., He, X., Kawaguchi, R. et al. Microglia-organized scar-free spinal cord repair in neonatal mice. Nature 587, 613–618 (2020). https://doi.org/10.1038/s41586-020-2795-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2795-6
This article is cited by
-
M2 microglia-derived exosome-loaded electroconductive hydrogel for enhancing neurological recovery after spinal cord injury
Journal of Nanobiotechnology (2024)
-
Fascin-1 limits myosin activity in microglia to control mechanical characterization of the injured spinal cord
Journal of Neuroinflammation (2024)
-
Animal models to study cardiac regeneration
Nature Reviews Cardiology (2024)
-
Endothelial cells and macrophages as allies in the healthy and diseased brain
Acta Neuropathologica (2024)
-
M2 Microglia-derived Exosomes Promote Spinal Cord Injury Recovery in Mice by Alleviating A1 Astrocyte Activation
Molecular Neurobiology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.