Abstract
Cell death in human diseases is often a consequence of disrupted cellular homeostasis. If cell death is prevented without restoring cellular homeostasis, it may lead to a persistent dysfunctional and pathological state. Although mechanisms of cell death have been thoroughly investigated1,2,3, it remains unclear how homeostasis can be restored after inhibition of cell death. Here we identify TRADD4,5,6, an adaptor protein, as a direct regulator of both cellular homeostasis and apoptosis. TRADD modulates cellular homeostasis by inhibiting K63-linked ubiquitination of beclin 1 mediated by TRAF2, cIAP1 and cIAP2, thereby reducing autophagy. TRADD deficiency inhibits RIPK1-dependent extrinsic apoptosis and proteasomal stress-induced intrinsic apoptosis. We also show that the small molecules ICCB-19 and Apt-1 bind to a pocket on the N-terminal TRAF2-binding domain of TRADD (TRADD-N), which interacts with the C-terminal domain (TRADD-C) and TRAF2 to modulate the ubiquitination of RIPK1 and beclin 1. Inhibition of TRADD by ICCB-19 or Apt-1 blocks apoptosis and restores cellular homeostasis by activating autophagy in cells with accumulated mutant tau, α-synuclein, or huntingtin. Treatment with Apt-1 restored proteostasis and inhibited cell death in a mouse model of proteinopathy induced by mutant tau(P301S). We conclude that pharmacological targeting of TRADD may represent a promising strategy for inhibiting cell death and restoring homeostasis to treat human diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The 3D atom coordinates of the TRADD–TRAF2 interaction complex were obtained from PDB (https://www.rcsb.org) with PDB ID 1F3V. All source data associated with the paper are provided as part of the paper. Source data for all figure plots are provided with the paper. The full gel blots and the proteomics datasets are provided in the Supplementary Information. Source data are provided with this paper.
References
Wallach, D., Kang, T. B., Dillon, C. P. & Green, D. R. Programmed necrosis in inflammation: Toward identification of the effector molecules. Science 352, aaf2154 (2016).
Shan, B., Pan, H., Najafov, A. & Yuan, J. Necroptosis in development and diseases. Genes Dev. 32, 327–340 (2018).
Ashkenazi, A. & Dixit, V. M. Death receptors: signaling and modulation. Science 281, 1305–1308 (1998).
Hsu, H., Xiong, J. & Goeddel, D. V. The TNF receptor 1-associated protein TRADD signals cell death and NF-κB activation. Cell 81, 495–504 (1995).
Pobezinskaya, Y. L. et al. The function of TRADD in signaling through tumor necrosis factor receptor 1 and TRIF-dependent Toll-like receptors. Nat. Immunol. 9, 1047–1054 (2008).
Hsu, H., Shu, H. B., Pan, M. G. & Goeddel, D. V. TRADD-TRAF2 and TRADD-FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84, 299–308 (1996).
Sweeney, P. et al. Protein misfolding in neurodegenerative diseases: implications and strategies. Transl. Neurodegener. 6, 6 (2017).
Morishita, H. & Mizushima, N. Diverse cellular roles of autophagy. Annu. Rev. Cell Dev. Biol. 35, 453–475 (2019).
Menzies, F. M. et al. Autophagy and neurodegeneration: pathogenic mechanisms and therapeutic opportunities. Neuron 93, 1015–1034 (2017).
Yoshii, S. R. & Mizushima, N. Monitoring and measuring autophagy. Int. J. Mol. Sci. 18, 1865 (2017).
Zhao, J., Zhai, B., Gygi, S. P. & Goldberg, A. L. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl Acad. Sci. USA 112, 15790–15797 (2015).
Noda, T. & Yoshimori, T. Molecular basis of canonical and bactericidal autophagy. Int. Immunol. 21, 1199–1204 (2009).
Zhong, Y. et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1–phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 (2009).
Matsunaga, K. et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 (2009).
Degterev, A. et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008).
Wertz, I. E. et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 528, 370–375 (2015).
Haas, T. L. et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009).
Ofengeim, D. & Yuan, J. Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 14, 727–736 (2013).
Vucic, D., Dixit, V. M. & Wertz, I. E. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat. Rev. Mol. Cell Biol. 12, 439–452 (2011).
Guo, X. et al. TAK1 regulates caspase 8 activation and necroptotic signaling via multiple cell death checkpoints. Cell Death Dis. 7, e2381 (2016).
Park, Y. C. et al. A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction. Cell 101, 777–787 (2000).
Shu, H. B., Takeuchi, M. & Goeddel, D. V. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc. Natl Acad. Sci. USA 93, 13973–13978 (1996).
Liu, J. et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell 147, 223–234 (2011).
Chen, N. J. et al. Beyond tumor necrosis factor receptor: TRADD signaling in toll-like receptors. Proc. Natl Acad. Sci. USA 105, 12429–12434 (2008).
Michallet, M. C. et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 28, 651–661 (2008).
Ermolaeva, M. A. et al. Function of TRADD in tumor necrosis factor receptor 1 signaling and in TRIF-dependent inflammatory responses. Nat. Immunol. 9, 1037–1046 (2008).
Callejas, N. A., Casado, M., Boscá, L. & Martín-Sanz, P. Requirement of nuclear factor kappaB for the constitutive expression of nitric oxide synthase-2 and cyclooxygenase-2 in rat trophoblasts. J. Cell Sci. 112, 3147–3155 (1999).
Apostol, B. L. et al. Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum. Mol. Genet. 15, 273–285 (2006).
Yoshiyama, Y. et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron 53, 337–351 (2007).
Iba, M. et al. Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J. Neurosci. 33, 1024–1037 (2013).
Jicha, G. A., Bowser, R., Kazam, I. G. & Davies, P. Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res. 48, 128–132 (1997).
Grøftehauge, M. K., Hajizadeh, N. R., Swann, M. J. & Pohl, E. Protein-ligand interactions investigated by thermal shift assays (TSA) and dual polarization interferometry (DPI). Acta Crystallogr. D 71, 36–44 (2015).
Cho, H. J. et al. Probing the effect of an inhibitor of an ATPase domain of Hsc70 on clathrin-mediated endocytosis. Mol. Biosyst. 11, 2763–2769 (2015).
Micheau, O. & Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003).
Anderton, H. et al. RIPK1 prevents TRADD-driven, but TNFR1 independent, apoptosis during development. Cell Death Differ. 26, 877–889 (2019).
Voortman, J., Checińska, A. & Giaccone, G. The proteasomal and apoptotic phenotype determine bortezomib sensitivity of non-small cell lung cancer cells. Mol. Cancer 6, 73 (2007).
Cox, J. & Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 (2008).
Acknowledgements
This work was supported in part by grants from UCB, FBRI-Fidelity, and the Quadrangle Fund for Advancing and Seeding Translational Research at Harvard Medical School (Blavatnik-Accelerator of Harvard and HMS/Q-FASTR) (to J.Y.). The work of D.X., L.Y., and B.S. during revision was supported in part by grants from the National Key R&D Program of China (2016YFA0501900), the China National Natural Science Foundation (31530041, 91849204 and 21837004), the Chinese Academy of Sciences (XDB39030000 and XDB39030600) and Shanghai Science and Technology Development Funds (20QA1411500). We thank L. Thompson for HTT(Q103)-PC12 cells; Z. Liu for Tradd−/− MEFs; V. Dixit for M1 and K63 ubiquitin antibodies; P. Davies for the MC1 antibody; G. Cuny for advice on chemistry; Z.-F. Wang and G. Heffron for advice on SPR and NMR; J. Smith for help with compound screening; J. Waters for help with microscopy; the EQNMR facility at HMS for their assistance; and C. Wang and C. Cao for the use of BIAcore.
Author information
Authors and Affiliations
Contributions
This project was conceived, designed, and directed by J.Y. D.X. and H. Zhao designed and conducted most of the experiments. D.X. and H.Z. made equal contributions as joint first authors and are listed alphabetically in the author list. M.J., H. Zhu, J.G. and S.A.D. conducted the multiplexed small molecule screen and contributed to early study of the compounds. Z.J.S. and G.W. assisted with NMR analysis of the TRADD–Apt-1 complex. J.X. conducted computational analysis of the TRADD–Apt-1 complex. Z.H. and V.M.Y.L. directed the tau transgenic mouse model study. Y.Q. and L.T. synthesized ICCB-19 and ICCB-19i for revision. B.S., P.A., L.M., M.G.N., A.N., L.Y., J.L., X.H., H.W., M.Z. and V.J.M. conducted some of the experiments. The manuscript was written by J.Y., D.X. and H. Zhao.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. Harvard University has filed a patent for the ICCB-19 and Apt-1 series of small molecules.
Additional information
Peer review information Nature thanks Kalle Gehring, Ingrid Wertz and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Isolation of ICCB-19 and Apt-1 for modulating both apoptosis and autophagy.
a, A multiplex chemical screening scheme for compounds that can modulate cellular homeostasis by activating autophagy and also block apoptosis. Primary screen: Jurkat cells were treated with Velcade (50 nM) and individual compounds (10 μM) in the library for 25h and cell viability was measured. 710 compounds which could protect against Velcade-induced apoptosis were selected. Secondary counterscreen: HCT116 cells were treated with 5-fluorouracil (5-FU) (100 μM) and individual compounds selected from the Primary screen (10 μM) for 24 h and cell viability was measured. The hits which protected against apoptosis induced by 5-FU were eliminated from further studies. Tertiary screen: H4-GFP-LC3 cells were treated with individual compounds (10 μM) for 24 h and GFP-LC3 dots were quantified. Quaternary screen: RGC-5 cells were treated with mTNF (0.5 ng/ml), TAK1 inhibitor (5Z)-7-Oxozeanol (0.5 μM) and individual compounds (10 μM) for 8 h and cell viability was measured. b, IC50s of ICCB-19 and Apt-1 protecting Velcade-induced apoptosis (50 nM) in Jurkat cells treated with indicated compounds for 24 h and cell viability was measured. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. c, IC50s of ICCB-19 and Apt-1 protecting RDA in MEFs were treated with mTNF (1 ng/mL) and 5Z-7-Oxozeaenol (0.5 μM) in the presence of indicated compounds at different concentrations for 8 h and cell survival was measured. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. d, KINOMEscan profiling of Apt-1 (10 μM) against a panel of 97 kinases. Binding interactions reported as % Ctrl, where lower numbers indicate stronger hits. Negative control = DMSO (100% Ctrl); positive control = control compound (0% Ctrl); 0 ≤ %Ctrl <0.1 Very Strong; 0.1 ≤ %Ctrl <1 Strong; 1 ≤ %Ctrl <10 Medium; 10 ≤ %Ctrl <35 Weak; %Ctrl ≥ 35 No effects. No significant binding of Apt-1 to this panel of 97 kinases was detected. e, H4 cells were treated with indicated concentrations of Apt-1. Autophagy was determined by LC3 II levels using immunoblotting. s.e. = shorter exposure, LE = longer exposure. Mean ± s.e.m. of n = 3 biologically independent experiments. f-i, SH-SY5Y (f), HeLa (g), HT-29 (h), and Jurkat (i) cells were treated with Apt-1 (10 μM), NH4Cl (20 mM) as indicated for 6 h. Autophagy was measured by LC3 II induction and p62 reduction by immunoblotting. The levels of LC3 II in cells treated with both Apt-1 and NH4Cl, the latter of which inhibits lysosome, were higher than that treated with either Apt-1 or NH4Cl alone. Thus, ICCB-19/Apt-1, but not ICCB-19i, induce autophagic flux. Mean ± s.e.m. of n = 3 independent experiments. Two-tailed t-test. **P = 0.0037 (f), 0.0024 (g), 0.0027 (h), 0.0036 (i). j, Effects of ICCB-19/Apt-1 on long-lived protein degradation. The rates of long-lived protein turnover in H4 cells treated with indicated compounds (10 μM, 6 h); rapamycin as positive control. Values expressed as fold changes relative to normal control cells. Mean ± s.e.m. of n = 4 biologically independent samples. One-way ANOVA, post hoc Dunnett’s test. k, MEFs and Jurkat cells treated with zVAD.fmk (20 μM) for 6 h.Levels of LC3 II determined by immunoblotting. l, MEFs were treated with vehicle (0 h), ICCB-19 (10 μM), or Apt-1 (10 μM) for indicated times. Cell lysates analysed by immunoblotting using indicated antibodies. m, H4-DsRed-FYVE cells were treated with indicated compounds for 6 h and imaged; representative cells shown. Average DsRed-FYVE puncta per 1000 cells from each sample was determined using ImageJ. Mean ± s.e.m. of the puncta per cell from n = 5 biologically independent experiments. One-way ANOVA, post hoc Dunnett’s test. **P = 0.0034; ***P = 0.0004; n.s. not significant (P = 0.6502). n, Beclin 1/Vps34 kinase complex isolated from Flag-Beclin 1 transfected HEK293T cells treated with ICCB-19, Apt-1, or ICCB-19i (10 μM) for 6 h. PI3P kinase activity was measured by in vitro lipid kinase assay using ADP-Glo Kinase Assay Kit. Wortmannin (10 μM) was used as a control to inhibit Vps34 kinase activity. Mean ± s.e.m. of n = 4 biologically independent samples. One-way ANOVA, post hoc Dunnett’s tests. ***P = 0.0003; 1 × 10−15 (left to right). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 cIAP1/2 and TRAF2 are required for induction of autophagy by ICCB-19/Apt-1.
a, HEK29T cells were transfected with Flag-Beclin 1 for 12 h, then treated with Apt-1 (10 μM) for another 12 h. Cell lysates were immunoprecipitated using anti-Flag beads. cIAP1 and TRAF2 levels were determined by immunoblotting. b, MEFs were treated with indicated concentrations of Apt-1 for 12 h. Cell lysates were immunoprecipitated using anti-Beclin 1 antibody. cIAP1 and TRAF2 levels were determined by immunoblotting. c, d, Long-lived protein turnover rates in MEFs with indicated genotypes treated with indicated compounds. Expressed as fold changes relative to normal control cells. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 3 × 10−14, 6 × 10−10, 1 × 10−7, 2 × 10−8 (left to right, c, d). e, MEFs were pre-treated with SM-164 (1 μM) for 1 h, then treated with Apt-1 for 6 h. LC3 II levels were determined by immunoblotting. Mean ± s.e.m. of n = 3 biologically independent experiments. Two-tailed t-test. ***P = 0.0003. f, shRNA-mediated TRAF2 stable knockdown MEFs were treated with Apt-1 (10 μM) for 6 h. LC3 II levels were determined by immunoblotting. Mean ± s.e.m. of n = 3 biologically independent experiments. Two-tailed t-test. **P = 0.0012. g, h, cIap1−/− and Traf2−/− MEFs reconstituted with HA-mcIAP1 and HA-mTRAF2, respectively, were treated with Apt-1 (10 μM) for 6 h. LC3 II levels were determined by immunoblotting. Mean ± s.e.m. of n = 3 biologically independent experiments. Two-tailed t-test**P = 0.0057 (g), ***P = 0.0007 (h). i, j, MEFs with indicated genotypes were treated with rapamycin (1 μM) for indicated time. LC3 II levels were determined by immunoblotting. The quantification of each experiment was shown on the right (n = 1), representative of 3 independent experiments. k, l, MEFs with indicated genotypes were incubated in HBSS for indicated time. LC3II levels were determined by immunoblotting. The quantification of each experiment was shown on the right (n = 1), representative of 3 independent experiments. m, MEFs were treated with indicated compounds for 6 h, then cell lysates were tandem-immunoprecipitated with anti-Beclin 1 antibody and denatured in 3 M urea. Anti-K63-linkage specific polyubiquitin antibody was used to conduct secondary immunoprecipitation. Samples were then immunoblotted with anti-Beclin 1 antibody to measure the K63-linkage specific ubiquitination of Beclin 1. n, MEFs were pretreated with SM-164 (1 μM) for 1 h, then treated with Apt-1 (10 μM) for 6 h, then K63-linkage specific ubiquitination of Beclin 1 was analysed as in (m). o, p, Reconstituted MEFs were treated with Apt-1 (10 μM) for 6 h, then K63-linkage specific ubiquitination of Beclin 1 was analysed as in (m). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 3 cIAP1 mediates K63-linked ubiquitination of Beclin 1 at K183 and K204.
a, Schematic representation of mass spectrometry assay to determine K63-linked ubiquitination sites of Beclin 1 by cIAP1. b, The quantitative mass spec analysis of K63-linked ubiquitination of each lysine site. c, Sequence alignment of key ubiquitination sites (K) within Beclin 1 orthologues from different species. See supplementary Table 2 for details. d, HEK293T cells were transfected with indicated plasmids for 24 h. Cells were lysed in 6 M urea and lysates were subjected to pull-down with Ni2+ beads and analysed by immunoblotting with anti-Beclin 1 antibody to detect ubiquitylated Beclin 1. e, Validation of Beclin 1 expression in Beclin 1-silenced H4 cells. f, Control and Beclin 1-silenced H4 cells were treated with Apt-1 (10 μM) for 6 h. LC3 II levels were determined by immunoblotting. g, Beclin 1-silenced H4 cells reconstituted with WT and mutants Beclin 1 were treated with Apt-1 (10 μM) for 6 h. LC3 II levels were determined by immunoblotting. Mean ± s.e.m. are quantified from n = 3 biologically independent experiments in graphs. Two-tailed t-test. **P = 0.0022 (d), 0.0049 (f), 0.0024 (g); *P = 0.0309, 0.0195 (left to right, d), 0.0126 (g); n.s. not significant, (P = 0.6959) (f). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 ICCB-19/Apt-1 block Velcade-induced apoptosis, RIPK1-dependent apoptosis and necroptosis.
a, b, Jurkat (a) or SH-SY5Y (b) cells were stimulated by Velcade (50 nM) in the presence of Apt-1 (10 μM), Nec-1 s (10 μM), or zVAD (20 μM) for 12 h and 24 h. The levels of cleaved caspase-3 were determined by immunoblotting. c, Tak1−/− MEFs were treated with 1 ng/ml mTNF in the presence of indicated compounds for 3 h. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. One-way ANOVA, post hoc Dunnett’s tests. ***P = 1 × 10−15 (left to right); n.s. not significant, (P = 0.7989). d, Tak1−/− MEFs were treated as in (a), the cell lysates were analysed by immunoblotting using indicated antibodies. e, MEFs were treated with mTNF (1 ng/ml) and 5Z-7-Oxozeaenol (0.5 μM) in the presence of indicated compounds for 1 h and 2 h and the cell lysates were analysed by immunoblotting using indicated antibodies. f, g, ICCB-19/Apt-1 inhibit RDA, including complex IIa formation (f) and caspase-8 activation (g). MEFs treated as in (e) were lysed with IP buffer and FADD was immunoprecipitated by anti-FADD antibody. Total lysates and IP samples were analysed by immunoblotting to determine the recruitment of RIPK1 to FADD in complex IIa (f). MEFs were treated with mTNF (1 ng/ml) and 5Z-7-Oxozeaenol (0.5 μM) in the presence of ICCB-19 (10 μM), Apt-1 (10 μM), Nec-1 s (10 μM), or zVAD.fmk (20 μM) for 4 h and the activity of caspase-8 was determined using Caspase-Glo 8 Assay Systems (g). Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. One-way ANOVA, post hoc Dunnett’s tests. ***P = 1 × 10−15 (g). h-k, RDA was induced in Tbk1−/− MEFs (h, i), Nemo−/− MEFs (j, k) by the treatment with mTNF (10 ng/ml) together with ICCB-19 (10 μM) and Nec-1 s (10 μM) at indicated times and cell death was determined by SYTOX Green (h, j) and caspase-3 cleavage (CC3) immunoblotting (i, k). Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments (h, j).). Two-way ANOVA. ***P = 5 × 10−7 (h), 4 × 10−7 (j). l, RDA was induced in WT MEFs by the treatment with mTNF (10 ng/ml) and IKK inhibitor TPCA-1 (5 μM) in the presence of ICCB-19 (10 μM) or Nec-1 s (10 μM) for indicated times and cell death was determined by SYTOX Green. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA. ***P = 1 × 10−5. m, Recombinant active caspase-8 was incubated with vehicle, ICCB-19 (10 μM), Apt-1 (10 μM), or zVAD.fmk (20 μM) for 1h and the activity of caspase-8 was determined using Caspase-Glo 8 Assay Systems. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. One-way ANOVA, post hoc Dunnett’s tests. n.s. not significant, (P = 0.9931, 0.9215 (left to right)). n, WT MEFs were treated with mTNF (1 ng/ml) and cycloheximide (CHX, 1 μg/mL) to induce RIA in the presence or absence of ICCB-19 (10 μM) or Nec-1 s (10 μM) for indicated time and cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 8 biologically independent samples, representative of 3 independent experiments. One-way ANOVA, post hoc Dunnett’s tests. n.s. not significant, (P = 0.9962). o, p65/p50 DKO MEFs were treated with mTNF (1 ng/ml) together with ICCB-19 (10 μM) for indicated times and cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA. n.s. not significant, (P = 0.1895). p, MEFs were treated as indicated and the cell survival was measured by CellTiter-Glo assay. The concentrations of reagents used: mTNF: 1 ng/mL; (5Z)-7-oxozeaenol: 0.5 μM; zVAD: 20 μM; ICCB-19: 10 μM; Apt-1: 10 μM; Nec-1 s: 10 μM. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. q, Necroptosis of MEFs was induced by the treatment with TNF/5z7/zVAD in the presence of indicated compounds for indicated hours and the activation of RIPK1(p-S166), RIPK3(p-T231/S232), and MLKL(p-S345) was determined by immunoblotting. r, HEK293T cells were transfected with Flag-RIPK1 expression construct for 12 h in the presence of Nec-1 s (10 μM), ICCB-19 (10 μM), or Apt-1 (10 μM). The activation of RIPK1 was determined by immunoblotting using p-RIPK1(S166) antibody. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 The protection of RDA by ICCB-19/Apt-1 requires TRADD.
a, b, Mass spectrometry analysis shown in Fig. 2b using ICCB-19 (a) or Apt-1 (b) were confirmed by immunoprecipitation-immunoblotting using indicated antibodies, quantified on the right (n = 1), representative of 3 independent experiments. c, MEFs were treated with Flag-mTNF (50 ng/ml) in the presence of Apt-1 (10 μM) for indicated time. The complex I was isolated by anti-Flag beads and denatured in 6 M urea. The complex I was further analysed by immunoprecipitation using anti-M1 (6 M urea) or K63 (3 M urea) ubiquitin antibody under denatured condition. The levels of RIPK1 ubiquitination were analysed by immunoblotting. d, WT and Traf2−/− MEFs were stimulated by mTNF (1 ng/ml) and 5Z-7-Oxozeaenol (0.5 μM) in the presence of indicated compounds for 8 h. Cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 0.0001. e, WT and cIap1/2−/− MEFs were stimulated by mTNF (10 ng/ml) in the presence of vehicle, ICCB-19 (10 μM) or Nec-1 s (10 μM) for indicated time. Cell death was determined by CellTiter-Glo assay. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA. f, MEFs were pretreated with SM-164 (50 nM) for 1 h, then stimulated by mTNF (10 ng/ml) in the presence of vehicle, ICCB-19 (10 μM) or Nec-1 s (10 μM) for indicated time. Cell death was determined by SYTOX Green assay. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA. ***P = 4 × 10−5; n.s. not significant, (P = 0.1772). g, cIap1/2−/− MEFs were stimulated with Flag-TNF (50 ng/ml) for indicated minutes in the presence of vehicle or ICCB-19 (10 μM) and the complex I was pulled down using anti-Flag beads. The levels of activated RIPK1 and total RIPK1 were determined by immunoblotting. h, i, cIap1/2−/− MEFs (h) and cIAP1-reconstituted cIAP1/2 DKO MEFs (i) were stimulated with Flag-TNF (50 ng/ml) for indicated minutes in the presence of vehicle or Apt-1 (10 μM) and the complex I was pulled down using anti-Flag beads. TRADD recruitment to complex I was determined by immunoblotting, quantified on the right (n = 1), representative of 3 independent experiments. j, k, Fadd-deficient and Ripk1-deficient Jurkat cells were treated with Velcade (50nM) in the presence of ICCB-19 (10 μM), Nec-1 s (10 μM), NAC (100 μM), or zVAD.fmk (20 μM). The activation of caspase-8, PARP cleavage (j), and activation of caspase-3 (k) were determined by immunoblotting. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 6 Autophagy is required for ICCB-19/Apt-1 to inhibit Velcade-induced apoptosis.
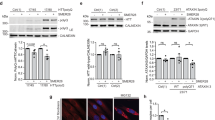
a, Tradd+/+ and Tradd−/− MEFs were treated with vehicle or ICCB-19 (10 μM) for 6 h. Autophagy levels were determined by immunoblotting using anti-LC3 antibody. Mean ± s.e.m. of n = 3 biologically independent experiments (right). Two-tailed t-test. n.s. not significant, (P = 0.9172). b, Tradd+/+ and Tradd−/− MEFs were treated with Apt-1 (10 μM), NH4Cl (10 mM) as indicated for 6 h. Autophagy levels were determined by immunoblotting using anti-LC3 antibody. Mean ± s.e.m. of n = 3 biologically independent experiments. Two-tailed t-test. n.s. not significant (P = 0.4064, 0.8913 (left to right)). c, Long-lived protein turnover rates in Tradd+/+ and Tradd−/− MEFs. Expressed as as fold changes relative to Tradd+/+ cells. Mean ± s.e.m. of n = 5 biologically independent samples, representative of 3 independent experiments. Two-tailed t-test. ***P = 0.0001. d, WT and Tradd-KO Jurkat cells were treated with Apt-1 (10 μM) and Spautin-1 (10 μM) followed by Velcade (50 nM) for 24 h. Cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 2 × 10−5, 3 × 10−15 (left to right). e, Jurkat cells were treated with ICCB-19 (10 μM), Apt-1 (10 μM), Chloroquine (50 μM), E64d (5 μg/ml) followed by Velcade (50 nM) for 24 h. The cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 4 × 10−14, 1 × 10−15 (left to right). f, Atg5-WT and Atg5-KO Jurkat cells were pretreated with Apt-1 (10 μM) or zVAD (20 μM) for 1 h, then stimulated by Velcade (50 μM) for 24 h. Cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 1 × 10−15. Validation of Atg5 knockout was determined by immunoblotting and shown on the right. g, Atg5+/+ and Atg5−/− MEFs were stimulated by TNF (1 ng/ml) and 5z7 (0.5 μM) for 8 h in the presence or absence of Apt-1 (10 μM). Cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 4 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. n.s. not significant, (P = 0.2568, 0.0822 (left to right)). h, HEK293T cells were transfected with indicated expression plasmids for 24 h. The whole-cell lysate lysed in 6 M urea was subjected to pull-down with Ni2+ beads and analysed by immunoblotting with anti-Beclin 1 antibody to detect ubiquitylated Beclin 1. The ubiquitination of Beclin 1 by cIAP1 was reduced upon overexpression of TRADD, which was restored by Apt-1. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 7 ICCB-19/Apt-1 inhibit inflammatory responses.
a, MEFs were stimulated by mTNF (10 ng/ml) in the presence of vehicle or ICCB-19 (10 μM) for indicated time. NF-κB and MAPKs activity were determined by immunoblotting using indicated abs. b, MEFs were stimulated by mTNF (10 ng/ml) in the presence of vehicle or ICCB-19 (10 μM) for indicated time. The protein levels of iNOS and Cox2 were determined by immunoblotting. c-g, BV2 cells (a microglial-like cell line) (c, d, e) and BMDMs (bone marrow-derived macrophages) (f, g) were treated with IFNγ (c, 1 unit/μl), Pam3CSK4 (ligand for TLR2) (e, 10 ng/ml), LPS (ligand for TLR4) (f, 10 ng/ml), or MDP (ligand for NOD2/RIPK2 pathway) (d, g, 10 μg/ml) for indicated time. The mRNA levels of TNF were determined using quantitative PCR. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments (c-g). h, BV2 cells were treated with IFNγ (1 unit/μl) together with Apt-1 (10 μM) or Nec-1 s (10 μM) for 24 h. TNF production was determined by ELISA. i, BV2 cells were pretreated with Apt-1 (10 μM) or Nec-1 s (10 μM) for 1 h and then MDP (10 μg/ml) was added to cells together with transfection reagent for 7 h. TNF production was determined by ELISA. j, BV2 cells were pretreated with Apt-1 (10 μM) or Nec-1 s (10 μM) and then treated with Pam3CSK4 (10 ng/ml) for 8 h. TNF production was determined by ELISA. k, BMDMs were pretreated with Apt-1 (10 μM) or Nec-1 s (10 μM) for 1h and then treated with LPS (10 ng/ml) for 7 h. TNF production was determined by ELISA. l, BMDMs were first primed with IFNγ (10 ng/ml) for 2 h and then removed. The cells were then treated with Apt-1 (10 μM) or Nec-1 s (10 μM) for 1 h and MDP (10 μg/ml) was added to cells directly and treated for 8 h. TNF production was determined by ELISA. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments (h-l). m, BMDMs were treated with LPS (10 ng/ml) in the presence of vehicle control or Apt-1 (10 μM) or Nec-1 s (10 μM) for indicated time. NF-κB and MAPKs activity were determined by immunoblotting using indicated abs. n, Body temperature was measured in male mice (n = 10, 8 weeks) injected with mTNF (9.5 μg, i.v.) after pretreatment with Apt-1 (20 mg/kg) 30 min before. Control mice (n = 9) received an equal amount of vehicle before mTNF challenge. Mean ± s.e.m. Two-way ANOVA. ***P = 0.0007. o, Kaplan–Meier Survival Curve was measured on mice treated as in (n). Two-sided log-rank (Mantel-Cox) test. ***P = 1 × 10−5. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 8 Restoring proteome homeostasis by ICCB-19/Apt-1.
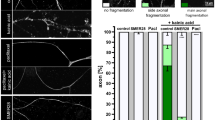
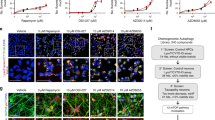
a, b, Parallel wells of PC12/Htt-Q103 cells were cultured with Vehicle, ICCB-19 (10 μM), ICCB-19i (10 μM), Apt-1 (10 μM), Nec-1 s (10 μM), and zVAD (20 μM) as indicated before the addition of Ponasterone A (5 μM) for 48 h. Nuclei were labelled with DAPI. The amount of Htt-Q103-EGFP aggregates per mm2 was quantified using ImageJ. Cell viability was measured by CellTiter-Glo assay. Survival rate is compared with vehicle-treated cells (b). Mean ± s.d. of n = 3 (a), n = 4 (b) biologically independent samples, representative of 3 independent experiments. One-way ANOVA, post hoc Dunnett’s tests. ***P = 1 × 10−5 (left to right, a, b); n.s. not significant, (P = 0.8556, 0.9195, 0.8613, 0.5687, 0.5304, 0.9998, 0.6029, 0.2821 (left to right, a, b)). c, d, SH-SY5Y cells were transfected with expression vectors for RFP-α-Synuclein WT, E46K, or A53T for 24h and then treated with vehicle or Apt-1 (10 μM) for 24 h. RFP-α-Synuclein was quantified by Fluorescence/Cell (RLU) by ImageJ. The cells treated as in (c) were lysed and analysed by immunoblotting for the levels of α-Synuclein (d). Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 8 × 10−5; *P = 0.0262, 0.0367 (left to right, c). e, f, Quantification of GFP-Tau fluorescence (e) or immunoblots of tau levels (f) in H4 cells treated with Apt-1. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. Two-way ANOVA, post hoc Bonferroni’s tests. ***P = 8 × 10−5, 8 × 10−4 (e, left to right). g, Representative images and quantification of cultured brain slices from PS19 mice (4 months old) stained with phospho-tau (red) and cell nuclei (DAPI/blue). Mean ± s.e.m. of n = 5 biologically independent samples. Two-tailed t-test. h, Immunoblots of tau levels in cultured PS19 mouse (4 months old) brain slices treated with indicated compounds. i, Pharmacokinetics of Apt-1 over 24 h dosing period in cerebrospinal fluid (CSF) and hippocampus. Apt-1 was delivered using intracerebroventricular Alzet micro-osmotic pump (20 mM Apt-1, 100 μl, release rate: 0.25 μl/h). CSF was collected at 1 h, 6 h, and 24 h. Hippocampi were collected at 24 h. The concentrations of Apt-1 were measured by HPLC. The concentration of Apt-1 in hippocampus at 24 h was 6.27 μM. Mean ± s.e.m. (n = 3 mice in each group). j, k, Synthetic preformed fibrils (pffs) [5μg full length tau (2N4R) with P301S mutation (T40/PS) per injection] or vehicle were injected into the hippocampi of PS19 mice (8 weeks old). Three weeks after the pffs injection, Apt-1 was delivered intracerebroventrically by Alzet micro-osmotic pumps (20 mM Apt-1, release rate 0.25μl/h) for one week before sacrificing. The hippocampi were isolated from the mice for immunoblotting using TAU-5 (Thermo Fisher) (j), or immunostaining for phospho-Tau (AT8) (k). Dots represent the mean from individual mice. Mean ± s.e.m. (n = 3 mice in each group). Two-tailed t-test. ***P = 0.0007. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 ICCB-19/Apt-1 reduce the interaction of TRADD and TRAF2.
a, Expression constructs encoding Flag-TRADD-N (1-179) and HA-TRADD-C (180-312) were transfected into HEK293T cells for 18 h. Then the cells were treated with Apt-1 (10 μM) or vehicle for another 6 h. The binding between Flag-TRADD-N (1-179) and HA-TRADD-C (180-312) was analysed by Co-IP assay as indicated. b-d, The effect of Apt-1 and ICCB-19 on the binding between TRADD-N and TRADD-C was determined by NanoBiT assay. Constructs were made encoding LgBiT and SmBiT fused to the N and C termini of TRADD-N and TRADD-C, respectively. HEK293T cells were transfected with these two plasmids for 24 h and then treated with indicated compounds for 6 h. The luminescence indicating the interaction of TRADD-N and TRADD-C was measured using Nano-Glo Live Cell Reagent. e, Test of the Nano-Bit system assay using a known binding pair: PRKAR2A and PRKACA. f, Apt-1 (10 μM) does not affect the Nano-Bit system assay as determined by using the known binding pair: PRKAR2A and PRKACA. g, Apt-1 reduces the binding between TRADD-N and TRAF2-C in a dose-dependent manner as determined by NanoBiT assay. h, Schematic representation of a cell-free Förster resonance energy transfer (FRET)-based assay to detect TRADDN and TRAF2C interaction. i, Purification of indicated proteins for FRET assay expressed in HEK293T cells. Proteins were pulled down by anti-Flag affinity gel and eluted by 3 × Flag peptide. CBB staining of the proteins are shown on the right. j, FRET-based assay to measure the direct interaction of TRADD-N and TRAF2-C was developed in which the donor Flag-TRAF2C-mCenulean (TRAF2C-mC) was excited at 430 nm, and the emission was measured from 450 to 600 nm. When FRET occurs, the acceptor mVenus-TRADDN-Flag (mV-TRADDN) emission will increase and the donor emission will decrease. Apt-1 was added to the system with indicated concentration and incubated for 1h, then subjected to FRET assay. k, Effect of TRAF2 on the binding between TRADD-N and TRADD-C was determined by NanoBiT assay. HEK293T cells were transfected with the plasmids as indicated for 24h and then treated with Apt-1 (10μM) for 4h, then luminescence was measured. l, U937 cells were stimulated with TNF (10 ng/ml) for indicated minutes in the presence of vehicle or Apt-1 (10 μM) and the complex I was pulled down using anti-TNFR1. Levels of cIAP1, TRAF2, and TRADD recruitment were determined by immunoblotting. Quantifications of cIAP1, TRAF2, and TRADD are shown below each blot. m, Due to the lack of a good anti-TRADD antibody for immunoprecipitation, Tradd−/− MEFs were reconstituted with Flag-mTRADD. Cells were treated with indicated concentrations of Apt-1 for 12 h, then co-IP was performed using anti-Flag antibody followed by immunoblotting using indicated antibodies. Mean ± s.d. of n = 3 (e, f), n = 4 (c, g), n = 5 (b), n = 6 (d, k) biologically independent samples, representative of 3 independent experiments. One-way ANOVA, post hoc Dunnett’s test. ***P = 1 × 10−15 (b, left to right), ***P = 1 × 10−15, n.s. not significant, (P = 0.3357) (d). ***P = 1 × 10−15 (k, left to right). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 10 ICCB-19/Apt-1 bind to TRADD-N.
A fluorescence-based thermal shift assay was developed to quantify ICCB-19/Apt-1 binding to TRADD by measuring changes in thermal denaturation temperature (Tm). a, CBB staining of GST-tag and GST-TRADD purified from HEK293T cells. b, in vitro binding of GST-TRADD (50 μM) with ICCB-19 (250 μM) and Apt-1 (250 μM) was determined by thermal shift assay. Thermal unfolding of GST-TRADD is monitored using SYPRO Orange. Data were collected in the presence of ICCB-19 and Apt-1, leading to a rightward shift in the unfolding transition. The apparent melting temperature (Tm) is the peak in the derivative of the unfolding curve (dF/dT), which is used as an indicator of thermal stability. c, GST-tag (50 μM) does not bind to the compounds (250 μM) as determined by thermal shift assay. d, ICCB-19i does not bind to GST-TRADD as determined by thermal shift assay. e, GST-TRADD-C (50 μM) alone does not bind to either ICCB-19 (250 μM) or Apt-1 (250 μM) as determined by thermal shift assay. f-h, TRADD-N/ICCB-19 (f), TRADD-N/Apt-1 (g), and TRADD-N/ICCB-19i (h) samples used for STD NMR experiments were prepared as 1 mM ICCB-19 (f), 1 mM Apt-1 (g), 1 mM ICCB-19i (h), and 13 μM TRADD-N in 0.5 mL of PBS in D2O (10%). The on-resonance irradiation of TRADD-N was performed at a chemical shift of -0.5 ppm, whereas the off-resonance irradiation was conducted at 37 ppm. Spectra were acquired using the following parameters: spectral window of 6.4 kHz, number of scans at 320, acquisition time of 2 s, and repetition time of 3 s. The decrease in signal intensity in STD spectrum, resulting from the transfer of saturation from the protein to the ligand, is evaluated by subtracting the on-resonance spectrum from the off-resonance spectrum. This subtraction yields a positive signal from a bound ligand. The asterisks indicate the signals of the compounds. The STD data suggest that both ICCB-19/Apt-1, but not ICCB-19i, bind with TRADD-N. i, CBB staining of 6 × His- and Flag-tagged TRADD for SPR purified from HEK293T cells. The proteins were pulled down by anti-Flag affinity gel and eluted by 3 × Flag peptide. The proteins were further purified by size exclusion chromatograph on a Superdex 75 column (GE Healthcare) in a buffer containing 20 mM imidazole (pH 6.6), 200 mM NaCl, 20 mM DTT. j, BIAcore SPR analysis of ICCB-19 binding to TRADD-N. The kinetic profile of ICCB-19 binding to TRADD-N is shown. A series of concentrations of ICCB-19 (ranging from 0.3125 to 10 μM) was used to measure the binding kinetics, with TRADD-N immobilized on the CM5 chip.
Extended Data Fig. 11 The binding mechanism of ICCB-19/Apt-1 on TRADD.
a-c, Superposition of 2D 1H-15N HSQC spectra of 15N-labelled His-TRADD-N (250 μM) in the presence (red) and absence (blue) of Apt-1 (500 μM) (a), ICCB-19 (500 μM) (b) or ICCB-19i (500 μM) (c). The close-up view of the region exhibited large perturbations was shown right. d, Binding pose of ICCB-19 in complex with TRADD-N was generated by induced-fit docking. The left panel demonstrated the shape and polarity of the ligand binding pocket surface, with red regions indicating negatively charged and blue positively charged. The right panel showed details of the interactions between the compound and TRADD-N. The compound was shown as cyan sticks, and the protein was shown as pink cartoon with key residues highlighted in sticks. Hydrogen bonds were shown as red dashed lines. e, Coomassie blue staining of WT and each mutant protein for thermal shift assay. f, g, HEK293T cells were seeded at 7.5 × 103 cells per well in a white, clear-bottom 96-well plate 24 h before transfection. Cells were then transfected with the indicated plasmids for 24h. Medium was removed and replaced with Opti-MEM medium (100 μl) for 1 h at 37 °C. The Nano-Glo reagent was prepared and added to each well immediately before the luminescence reading was taken. Luminescence was measured immediately on a plate reader and reported as relative light units (RLU). Mean ± s.d. of n = 6 biologically independent samples, representative of 3 independent experiments.
Extended Data Fig. 12 A mutagenesis study of ICCB-19/Apt-1 binding with TRADD to inhibit RDA and activate autophagy.
a, Tradd−/− MEFs were reconstituted with Flag-tagged WT or mutant TRADD as indicated. Expression levels of TRADD were determined by immunoblotting. b, Tradd−/− MEFs transfected with Flag-tagged WT or indicated TRADD mutants were stimulated by TNF/5z7 for 9 h in the presence or absence of Apt-1 (10 μM). Cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. Two-tailed t-test. c, d, TRADD-N(G121A)/Apt-1 (c), and TRADD-N(G121A)/ICCB-19 (d) samples for STD-NMR analyses were prepared as that of WT TRADD in (10f) with 1 mM Apt-1 (c), 1 mM ICCB-19 (d), and 13 μM TRADD-N(G121A) in 0.5 mL of PBS in D2O (10%). e, BIAcore SPR analysis of Apt-1 binding to TRADD-N(G121A). The kinetic profile of Apt-1 binding to TRADD-N(G121A) is shown. A series of concentrations of Apt-1 (ranging from 0.15625 to 5 μM) was used to measure the binding kinetics, with TRADD-N(G121A) immobilized on the CM5 chip. f, Tradd−/− MEFs were reconstituted with Flag-tagged WT or indicated TRADD mutants. The expression levels of TRADD were determined by immunoblotting. g, h, Tradd−/− MEFs reconstituted with Flag-mutant TRADD (Y16A/F18A or Y16A/I72A/R119A) were stimulated by TNF/5z7 for indicated time in the presence or absence of Apt-1 (10 μM). The cell survival was determined by CellTiter-Glo assay. Mean ± s.d. of n = 3 biologically independent samples, representative of 3 independent experiments. i, A model for mechanism by which Apt-1 targets TRADD to inhibit RDA and activate autophagy. In TNF-stimulated cells: Apt-1 binds to TRADD-N to reduce its binding with TRADD-C which stabilizes the binding of TRADD mediated by its DD in TRADD-C with the DD in TNFR1. The binding of Apt-1 with TRADD in complex I modulates the K63/M1-linked ubiquitination of RIPK1 by reducing the binding of TRADD with TRAF2/cIAP1/2 which increases the recruitment of A20 and HOIP to inhibit the activation of RIPK1 kinase. Increased retention of TRADD in complex I also decreases cytosolic availability of TRADD for the formation of complex IIa in which TRADD is known to be a key component. Treatment with Apt-1 also reduces the activation of NF-κB in TNF-stimulated cells. In autophagy pathway: TRADD normally binds to TRAF2 and cIAP1/2 homeostatically. Apt-1 can release TRAF2 and cIAP1/2 from their binding with TRADD. Released TRAF2/cIAP1/2 in turn mediates K63-linked ubiquitination of Beclin 1 to promote the formation of Vps34 complex, production of PtdIns3P, and activation of autophagy. For gel source data, see Supplementary Fig. 1.
Supplementary information
Supplementary Figure
Supplementary Figure 1. Full scan of all gel blots. Full blot images of all cropped bands presented in data figures, unless shown elsewhere already. Note that all the loading controls were on the same gel as target protein.
Supplementary Figure
Supplementary Figure 2. Synthesis and chemical structure characterization of Apt-1. Synthesis of Apt-1 and chemical structure characterization of Apt-1, including 1H NMR spectra and LRMS spectra of Apt-1.
Supplementary Table
Supplementary Table 1. Proteomics analysis of Beclin 1-VPS34 complex. Related to Fig. 1c. An excel file of the proteomics data showing the quantification of the abundance of each protein interacted with Beclin 1 in the presence or absence of Apt-1.
Supplementary Table
Supplementary Table 2. Proteomics analysis to identify the ubiquitination sites of Beclin 1 mediated by cIAP1. Related to Extended Data Fig. 3b. The quantitative mass spec analysis of K63 ubiquitination of each lysine site of Beclin 1 in the presence or absence of cIAP1.
Supplementary Table
Supplementary Table 3. Proteomics analysis of TNF-RSC complex (complex I). Related to Fig. 2b. An excel file of the proteomics data showing the quantification of the abundance of each protein in complex I in the presence or absence of ICCB-19.
Source data
Rights and permissions
About this article
Cite this article
Xu, D., Zhao, H., Jin, M. et al. Modulating TRADD to restore cellular homeostasis and inhibit apoptosis. Nature 587, 133–138 (2020). https://doi.org/10.1038/s41586-020-2757-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2757-z
This article is cited by
-
NAP1L1 regulates BIRC2 ubiquitination modification via E3 ubiquitin ligase UBR4 and hence determines hepatocellular carcinoma progression
Cell Death Discovery (2024)
-
Deciphering a cell death-associated signature for predicting prognosis and response to immunotherapy in lung squamous cell carcinoma
Respiratory Research (2023)
-
TRAF2 promotes M2-polarized tumor-associated macrophage infiltration, angiogenesis and cancer progression by inhibiting autophagy in clear cell renal cell carcinoma
Journal of Experimental & Clinical Cancer Research (2023)
-
Inhibition of TRADD ameliorates chondrocyte necroptosis and osteoarthritis by blocking RIPK1-TAK1 pathway and restoring autophagy
Cell Death Discovery (2023)
-
A guide to cell death pathways
Nature Reviews Molecular Cell Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.