Abstract
The DNA sensor cyclic GMP–AMP synthase (cGAS) initiates innate immune responses following microbial infection, cellular stress and cancer1. Upon activation by double-stranded DNA, cytosolic cGAS produces 2′3′ cGMP–AMP, which triggers the induction of inflammatory cytokines and type I interferons 2,3,4,5,6,7. cGAS is also present inside the cell nucleus, which is replete with genomic DNA8, where chromatin has been implicated in restricting its enzymatic activity9. However, the structural basis for inhibition of cGAS by chromatin remains unknown. Here we present the cryo-electron microscopy structure of human cGAS bound to nucleosomes. cGAS makes extensive contacts with both the acidic patch of the histone H2A–H2B heterodimer and nucleosomal DNA. The structural and complementary biochemical analysis also find cGAS engaged to a second nucleosome in trans. Mechanistically, binding of the nucleosome locks cGAS into a monomeric state, in which steric hindrance suppresses spurious activation by genomic DNA. We find that mutations to the cGAS–acidic patch interface are sufficient to abolish the inhibitory effect of nucleosomes in vitro and to unleash the activity of cGAS on genomic DNA in living cells. Our work uncovers the structural basis of the interaction between cGAS and chromatin and details a mechanism that permits self–non-self discrimination of genomic DNA by cGAS.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The electron density reconstructions and corresponding final models for NCP1–cGAS1–cGAS2–NCP2 and NCP1–cGAS1 were deposited in the Electron Microscopy Data Bank (accession codes: EMDB-10694 and EMDB-10695) and in the PDB (accession codes: 6Y5D and 6Y5E). The electron density reconstructions for NCP1–WT cGAS1–WT cGAS2–NCP2 and NCP1–WT cGAS1 were deposited in the Electron Microscopy Data Bank (accession codes: EMDB-11006 and EMDB-11005).
References
Ablasser, A. & Chen, Z. J. cGAS in action: expanding roles in immunity and inflammation. Science 363, eaat8657 (2019).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Wu, J. et al. Cyclic GMP–AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Ablasser, A. et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature 498, 380–384 (2013).
Gao, P. et al. Cyclic [G(2′,5′)pA(3′,5′)p] is the metazoan second messenger produced by DNA-activated cyclic GMP–AMP synthase. Cell 153, 1094–1107 (2013).
Diner, E. J. et al. The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING. Cell Rep. 3, 1355–1361 (2013).
Zhang, X. et al. Cyclic GMP–AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013).
Gentili, M. et al. The N-terminal domain of cGAS determines preferential association with centromeric DNA and innate immune activation in the nucleus. Cell Rep. 26, 2377–2393.e13 (2019).
Volkman, H. E., Cambier, S., Gray, E. E. & Stetson, D. B. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. eLife 8, e47491 (2019).
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Orzalli, M. H. et al. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl Acad. Sci. USA 112, E1773–E1781 (2015).
Lahaye, X. et al. NONO detects the nuclear HIV capsid to promote cGAS-mediated innate immune activation. Cell 175, 488–501.e22 (2018).
Zierhut, C. et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315.e23 (2019).
Pang, B. et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat. Commun. 4, 1908 (2013).
Stark, H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 (2010).
Li, X. et al. Cyclic GMP–AMP synthase is activated by double-stranded DNA-induced oligomerization. Immunity 39, 1019–1031 (2013).
McGinty, R. K. & Tan, S. Nucleosome structure and function. Chem. Rev. 115, 2255–2273 (2015).
Civril, F. et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 498, 332–337 (2013).
Barbera, A. J. et al. The nucleosomal surface as a docking station for Kaposi’s sarcoma herpesvirus LANA. Science 311, 856–861 (2006).
Abe, T. & Barber, G. N. Cytosolic-DNA-mediated, STING-dependent proinflammatory gene induction necessitates canonical NF-κB activation through TBK1. J. Virol. 88, 5328–5341 (2014).
Zhang, X. et al. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 6, 421–430 (2014).
Andreeva, L. et al. cGAS senses long and HMGB/TFAM-bound U-turn DNA by forming protein–DNA ladders. Nature 549, 394–398 (2017).
Zhou, W. et al. Structure of the human cGAS–DNA complex reveals enhanced control of immune surveillance. Cell 174, 300–311.e11 (2018).
Xie, W. et al. Human cGAS catalytic domain has an additional DNA-binding interface that enhances enzymatic activity and liquid-phase condensation. Proc. Natl Acad. Sci. USA 116, 11946–11955 (2019).
Ablasser, A. et al. Cell intrinsic immunity spreads to bystander cells via the intercellular transfer of cGAMP. Nature 503, 530–534 (2013).
Dobbs, N. et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 (2015).
Konno, H., Konno, K. & Barber, G. N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013).
Janeway, C. A. Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989).
Ablasser, A. & Hur, S. Regulation of cGAS- and RLR-mediated immunity to nucleic acids. Nat. Immunol. 21, 17–29 (2020).
Guey, B. et al. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 369, 823–828 (2020).
Dolinsky, T.J. et al. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004).
Ran, F. A. et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protocols 8, 2281–2308 (2013).
Haag, S. M. et al. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018).
Marks, B. D. et al. Multiparameter analysis of a screen for progesterone receptor ligands: comparing fluorescence lifetime and fluorescence polarization measurements. Assay Drug Dev. Technol. 3, 613–622 (2005).
Hanson, B. L., Alexander, C., Harp, J. M. & Bunick, G. J. Preparation and crystallization of nucleosome core particle. Methods Enzymol. 375, 44–62 (2004).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304, 3–19 (1999).
Schenk, A. D., Cavadini, S., Thomä, N. H. & Genoud, C. Live analysis and reconstruction of single-particle cryo-electron microscopy data with CryoFLARE. J. Chem. Inf. Model. 60, 2561–2569 (2020).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
de la Rosa-Trevín, J. M. et al. Xmipp 3.0: an improved software suite for image processing in electron microscopy. J. Struct. Biol. 184, 321–328 (2013).
Matsumoto, S. et al. DNA damage detection in nucleosomes involves DNA register shifting. Nature 571, 79–84 (2019).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Terwilliger, T. C. et al. phenix.mr_rosetta: molecular replacement and model rebuilding with Phenix and Rosetta. J. Struct. Funct. Genomics 13, 81–90 (2012).
Lebedev, A. A. et al. JLigand: a graphical tool for the CCP4 template-restraint library. Acta Crystallogr. D Biol. Crystallogr. 68, 431–440 (2012).
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007).
Waterhouse, A. M., Procter, J. B., Martin, D. M., Clamp, M. & Barton, G. J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
Acknowledgements
We thank N. Jordan and J. Ricci for technical assistance. A.A. received grants from the SNF (BSSGI0-155984 and 31003A_159836), NCCR in Chemical Biology, and was supported by the European Union’s Horizon 2020 Research and Innovation program grant agreement (grant no: 804933, ImAgine). N.H.T. was supported by the European Research Council (ERC) under the European Union’s Horizon 2020 Research and Innovation program grant agreement (grant no. 666068), the Novartis Research Foundation and by the SNF (31003A_179541). B.F. is supported by the European Union’s Horizon 2020 Research and Innovation program grant agreement (grant no: 724022, chromo-SUMMIT) and the NCCR in Chemical Biology. Z.K. was supported by the European Union’s Horizon 2020 Research and Innovation program under the Marie Skłodowska-Curie grant agreement no. 765445.
Author information
Authors and Affiliations
Contributions
G.R.P. conducted the cryo-electron microscopy experiments and data processing with help from S.C. and G.K. for the model building. A.D. performed the in vitro assays and purified recombinant cGAS. S.G., P.M. and A.A. performed the cellular experiments. K.M. and R.H. purified recombinant histone proteins and assembled the recombinant nucleosomes and chromatin fibres. Electron microscopy samples were prepared by G.R.P. with help from J.W. Z.K. and J.W. performed the fluorescence polarization assays. B.G. performed the FRAP experiments. B.F. supervised experiments related to the in vitro reconstitution of nucleosomes/chromatin and provided valuable discussion. N.H.T. supervised the structural work and the biophysical assays. A.A. supervised the in vitro assays and cellular studies. N.H.T. and A.A. wrote the manuscript with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
A.A. is a member of the scientific advisory board of IFM Therapeutics and scientific co-founder of IFM-Due.
Additional information
Peer review information Nature thanks Osamu Nureki and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
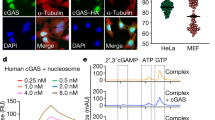
Extended Data Fig. 1 H2A-H2B dimers bind to and inhibit cGAS.
a, Confocal microscopy images of human BJ fibroblasts stained with primary antibodies against cGAS (green) and H2B (red). DNA was stained with DAPI (blue). Scale bar, 25 μm. b, Human BJ fibroblasts were treated with aclarubicin (20 μM) as indicated. Differential nuclear salt fractions were obtained, and the presence of the indicated proteins within each fraction was monitored by immunoblot. In a and b, the experiments were independently repeated at least three times. c, Human BJ fibroblasts were treated with DMSO (control) or aclarubicin (20 μM) for 2 h. After fixation, cells were subjected to PLA with anti-cGAS, anti-H2B, and anti-H4, respectively. PLA signals were quantified from at least 50 individual cells. Representative images are displayed (left) and data (right) are mean ± s.d. of one representative experiment out of n = 3 independent experiments. Two-tailed student`s t-test. Scale bar, 20 μm. d, Specificity control for PLA with human BJ fibroblasts using single antibody staining for cGAS, H2B, and H4, respectively. Scale bar, 20 μm. The experiment was repeated three times with similar results. e, Relative levels of in vitro cGAMP synthesis in the presence or absence of a concentration gradient of nucleosomes (from 75 nM to 1 nM) and chromatin fibres (from 6 nM to 0.1 nM). f, g, Relative levels of in vitro cGAMP synthesis in the presence or absence of a concentration gradient of histone H2A, H2B or H2A-H2B dimers (from 5 μM to 0.3125 μM; 1:2 step dilutions) (f) or H3 and H4 (from 5 μM to 0.3125 μM) (g). cGAS (catalytic domain; aa 155-522; hcGAS) activation was induced by HT DNA and data are mean ± s.d. of n = 3 independent experiments. One-way ANOVA with post hoc Dunnett multiple comparison test (e–g). h, Calculated IC50 values without (IC50) or with (cor. IC50) correction for the number of cGAS binding sites per molecule (nucleosome or fibre). Data points are from experimentally independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Cryo-EM analysis of cGAS bound to nucleosomes.
a, Cryo-EM micrograph of the non-crosslinked sample containing wild-type cGAS(WT) bound to NCP. Scale bar, 20 nm (Micrograph is representative of 20 images taken). b, Cryo-EM micrograph of the non-crosslinked sample containing dimerization mutant cGAS(K392E) bound to NCP. Scale bar, 20 nm (Micrograph is representative of 20 images taken). The cGAS-NCP complexes were directly concentrated and frozen on grids after gel filtration chromatography. Chromatin fibres induced by cGAS binding are highlighted by white rectangles (a, b). c, d, Magnified view of oligomeric assemblies extracted from a and b (starred rectangles), respectively.
Extended Data Fig. 3 Schematics of cGAS-NCP structure determination, classification and refinements.
a, Representative denoised cryo-EM micrograph of cGAS-NCP complex derived using JANNI (n = 2,890 micrographs). Scale bar, 20 nm. b, Reference-free 2D class averages of the particles picked using crYOLO. c, Data processing scheme starting with an ab initio model derived from 2D classes (b) for 34,000 particles. The scheme is divided into two separate data sets. On the right, data processing for in the a 4.1 Å dimeric structure of 2NCP-2cGAS. 103,688 particles were subjected to 3D classification using RELION 3.042 leading to four classes. The best class containing dimeric particles with 2NCP-2cGAS was further subjected to a second round of 3D classification producing two classes. The class containing 13,943 dimeric particles was further polished, 3D refined using RELION 3.0 and later CTF refined using cryoSPARC40, generating a 4.1 Å map. The left side of the scheme shows the data processing for the 3.1 Å structure of 1NCP-1cGAS. 289,518 particles were subjected to 3D classification using RELION 3.042 resulting in four classes. The best class containing 87,323 dimeric particles with 2NCP-2cGAS was further polished and merged with the 13,943 particles used for 4.1 Å map. The merged particles were subjected to CTF refinement and signal subtraction for the density accounting for 1cGAS-1NCP. 3D classification followed by non-uniform refinement in cryoSPARC led to a 3.1 Å map. d, Gold-standard Fourier shell correlation curves are shown for the 3.1 Å monomer map (blue) and the 4.1 Å dimer map (orange). e, f, Local resolution filtered maps (MonoRes) for the 4.1 Å dimer map and 3.1 Å monomer map, respectively. g, h, Angular distribution plots shown for the 3.1 Å monomer map and 4.1 Å dimer map respectively.
Extended Data Fig. 4 Cryo-EM structure of wild-type cGAS bound to nucleosomes.
a, A 5.1 Å 3D reconstruction of the complex containing two wild-type cGAS protomers, cGAS1 (red) and cGAS2 (orange), and two nucleosomal core particles, NCP1 and NCP2, respectively. b, Ribbon diagram of the dimerization mutant (K394E) NCP1-cGAS1-cGAS2-NCP2 model rigid-body fit into wild type NCP1-cGAS1-cGAS2-NCP2 electron-density maps. c, 3D focused classification map of NCP1-cGAS1 shown at low contour levels. d, Magnified view of cGAS1 and NCP1 DNA interactions. The zinc ion (cyan sphere) is coordinated by residues H390, C396, C397 and C404 forming the zinc finger motif. Residue K394 (sapphire blue sphere), is part of loop coordinating the zinc ion and is positioned close to the DNA of NCP1. Electron density connecting the NCP1 DNA and the cGAS1 zinc ion coordinating loop is shown in the background. e, A 4.1 Å EM map of the dimerization mutant cGAS(K394E)-NCP complex. f, A 5.1 Å EM map of the wild-type cGAS-NCP complex. g, EM maps from the mutant cGAS-NCP complex (grey) (e) superposed onto the wild-type cGAS-NCP complex map (blue) (f). The map-to-map fit gives a correlation value of 0.87 (Extended Data Table 1(b)).
Extended Data Fig. 5 Schematics of wild-type cGAS-NCP structure determination, classification and refinements.
a, Representative denoised cryo-EM micrograph of cGAS-NCP complex derived using JANNI (n = 5,007 micrographs). Scale bar, 25 nm. b, Reference-free 2D class averages of the particles picked using crYOLO. c, Data processing scheme starting with an ab initio model derived from 2Ds (b) is shown using 16,000 particles. The scheme is divided into two sub-schemes. The left side shows the processing of data for a 5.1 Å dimeric structure of NCP1-WTcGAS1-WTcGAS1-NCP1. 142,743 particles were subjected to 3D hetero refinement classification using cryoSPARC leading to two classes. The best class containing dimeric particles with WTcGAS2-NCP2 was further subjected to local refinement and later CTF refined using cryoSPARC generating a 5.1 Å map. For the scheme on the right, 56,747 particles were subjected to particle subtraction. The class containing 56,747 dimeric particles with WTcGAS1-NCP1 was locally refined and later CTF refined using cryoSPARC generating a 4.7 Å map. d, Gold-standard Fourier shell correlation curves are shown for the reconstructions, 5.1 Å (blue) and 4.7 Å (red). e, Local resolution filtered maps (MonoRes) for the 5.1 Å dimer map. f, Local resolution filtered maps (MonoRes) for the 4.7 Å monomer map. g, Angular distribution for the dimer 5.1 Å map. h, Angular distribution for the 4.7 Å monomer map.
Extended Data Fig. 6 Interaction of cGAS1 with NCP1 and mechanism of inhibition.
a, Ribbon diagram and a 3D reconstruction of the complex containing cGAS1 (red) and NCP1 (grey, blue). b–f, EM densities (shown as mesh at 4.5σ) for the residues interacting with the nucleosome in cis. Shown are cGAS Loop1, Loop2 and Loop3 interactions in cis with H2A-H2B dimer respectively (b–d) and the interactions of cGAS helix α5 residues with the DNA backbone from NCP1 (e, f). g, Overview of the hcGAS-DNA 2:2 minimal cGAS-DNA active dimer complex to highlight the two distinct DNA-binding surfaces (A-site and B-site) as previously defined16 (PDB: 4LEY). h, Superposition of the hcGAS-DNA complex (g) onto the cGAS-NCP complex as defined in this work (Fig. 2g). i, Magnified view of the common binding site (B-site) showing the clash of the two DNA strands with the nucleosomal DNA (orange, ligand DNA and grey, nucleosomal DNA).
Extended Data Fig. 7 Interaction of cGAS with nucleosomes in vitro.
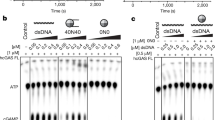
a, EMSA gel showing the interaction of nucleosomes (40 ng μl−1) with WT cGAS (40 pmol) in presence of increasing concentrations of LANA peptide WT or AA mutant (from 0.6 mg ml−1 to 78 μg ml−1; 1:2 step dilution). Dark grey arrowhead: nucleosomes complexed with cGAS, light grey arrowhead: free nucleosomes. Data are representative for three independent experiments. b, In vitro cGAMP synthesis of WT hcGAS (50 nM) with chromatin (5 nM) in presence of a gradient concentration of LANA peptide WT or AA mutated (from 0.5 mg ml−1 to 0.125 mg ml−1; 1:2 step dilution) normalized by cGAMP levels in absence of chromatin. Mean ± s.d. of n = 3 independent experiments is shown. One-way ANOVA with post hoc Dunnett multiple comparison test. Data points are from independent experiments. c, In vitro cGAMP synthesis of WT, R255A and R236A hcGAS (all 200 nM) with a concentration gradient of 147bp dsDNA or nucleosome (no DNA overhang) (left) or 227bp dsDNA or nucleosomes (80bp dsDNA overhang) (right) (from 200 nM to 50 nM; 1:2 step dilutions) normalized to cGAMP levels for 200 nM dsDNA for each individual mutant. Mean ± s.d. of n = 3 independent experiments is shown. d, Fluorescein (Flc) labelled 21 bp dsDNA tracer (10 nM) were mixed with cGAS protein (300 nM) and counter-titrated with unlabelled DNA (left) or nucleosomes (right) (see Methods). e, Forward titration experiments using 10 nM Flc-labelled probe in the presence of increasing amounts of either WT cGAS or cGAS pre-treated with 0.1-5 mM EDTA as indicated. f, Forward titration as in a but with cGAS K394E, either pre-treated with 0.1 mM EDTA or not. g, As in (d) but the cGAS protein was pre-treated with EDTA as indicated. For d and g, all data include two technical replicates and all data points are explicitly shown. Affinities are indicated as IC50 values. For e and f, all data include three technical replicates and are shown as mean ± s.d. Affinities are indicated as apparent Kd (Kapp) values. For gel source data, see Supplementary Fig. 1.
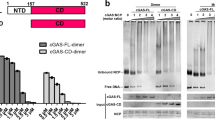
Extended Data Fig. 8 cGAS interactions with the second nucleosome in trans.
a, EM envelope of the dimeric complex containing NCP1-cGAS1-cGAS2-NCP2. b, A magnified view of an additional density accounting for N-terminal tail of histone H4. c, A 3D class showing the additional EM density found (dotted circles) near the outward facing acidic patch. The dimeric structure of NCP1-cGAS1-cGAS2-NCP2 is modelled into the major density. d, Modelling of cGAS molecules on the outer sides of the two NCP molecules in the map (c) resulting in a complex of cGAS1’-NCP1-cGAS1-cGAS2-NCP2-cGAS2’. e, A representative micrograph containing a cGAS (WT)-NCP multimeric complex (n = 20 micrographs). f, Magnified view on oligomeric assemblies extracted from (e) (left), Scale bar, 10 nm, and a modelled arrangement of cGAS-NCP in cartoon representation (right). g, Differential nuclear salt fractions probed for cGAS and H2B by immunoblot from HeLa cGAS KO cells reconstituted with doxycycline-inducible WT cGAS or cGAS K285A/R300A/K427A after 2 days of doxycycline treatment. One representative experiment of n = 3 independent experiments is shown. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 Effect of structure-guided mutations in cells.
a, Lines show FRAP recovery curves obtained after photo-bleaching WT cGAS-GFP or cGAS-GFP mutants inside the nucleus of cGAS KO HeLa cells. Data show mean ± SEM from 20-25 measurements. Graph is representative of n = 3 (left panel) or 4 (right panel) independent experiments. b–d, HeLa cGAS KO cells reconstituted with doxycycline-inducible WT cGAS or cGAS mutants were treated with doxycycline (1μg/ml) for 16h and 40h (b, c) or 40h (d), respectively. In b, cells were lysed and mRNA levels of IFI44, IFIT2 and CGAS were assessed. Data are presented as fold induction relative to non-treated WT cGAS and are mean ± s.d. of n = 5 independent experiments. Two-way ANOVA with post hoc Tukey multiple comparison test. In c, cells were lysed and STING and GAPDH levels were assessed by immunoblot. One representative experiment for n = 3 (16h) and n = 3 (40h) experiments with similar results is shown. In d, cells were stimulated with dsDNA (90mer) for 4h or left untreated (Ctrl.) and mRNA levels of IFI44 and IFIT2 were measured. Data show mean ± s.d. of n = 2 independent experiments. Individual data points represent biological replicates. For gel source data, see Supplementary Fig. 1. e, cGAS multiple sequence alignments showing the sequence conservation of residues involved in interactions with the acidic patch, nucleosome binding in cis, and nucleosome binding in trans. cGAS sequences from human, monkey, bovine, pig, mouse and rat corresponding to UniProt ID’s Q8N884, F7B8L6, E1BGN7, I3LM39, Q8C6L5 and A0A0G2JVC4 have been used in the alignment. The key residues involved in the interactions are highlighted in cyan. The consensus sequence and logo representation of the residues is shown below the sequence alignment. The alignment figure was created using Jalview52.
Supplementary information
Supplementary Figure
This file contains the uncropped blots.
Supplementary Video1
NCP1-cGAS1-cGAS2-NCP2 sandwich arrangement This file contains Supplementary Video1 which shows the surface and cartoon representation of the sandwich arrangement of NCP1-cGAS1-cGAS2-NCP2.
Supplementary Video2
NCP1-cGAS1 monomeric arrangement This file contains Supplementary Video2 which shows the surface and cartoon representation of monomeric arrangement of cGAS1 bound to NCP1.
Supplementary Video3
Structural insights into the cGAS1-NCP1 complex This file contains Supplementary Video3 which highlights the bipartite interactions of cGAS1 bound to NCP1 via Loop1, Loop2, Loo3 and the cGAS-DNA.
Supplementary Video4
Mechanism of cGAS inhibition by NCP1 This file contains Supplementary Video4 which shows how the cGAS activation is prevented by H2A-H2B dimer and also by nucleosome. Additionally, it highlights the steric clashes that occurs when a second cGAS1’ molecule would try to dimerize with cGAS1.
Rights and permissions
About this article
Cite this article
Pathare, G.R., Decout, A., Glück, S. et al. Structural mechanism of cGAS inhibition by the nucleosome. Nature 587, 668–672 (2020). https://doi.org/10.1038/s41586-020-2750-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2750-6
This article is cited by
-
The CRL5–SPSB3 ubiquitin ligase targets nuclear cGAS for degradation
Nature (2024)
-
DNA sensing and repair systems unexpectedly team up against cancer
Nature (2024)
-
TXNRD1 drives the innate immune response in senescent cells with implications for age-associated inflammation
Nature Aging (2024)
-
When DNA-damage responses meet innate and adaptive immunity
Cellular and Molecular Life Sciences (2024)
-
MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.