Abstract
Heterochromatin that depends on histone H3 lysine 9 methylation (H3K9me) renders embedded genes transcriptionally silent1,2,3. In the fission yeast Schizosaccharomyces pombe, H3K9me heterochromatin can be transmitted through cell division provided the counteracting demethylase Epe1 is absent4,5. Heterochromatin heritability might allow wild-type cells under certain conditions to acquire epimutations, which could influence phenotype through unstable gene silencing rather than DNA change6,7. Here we show that heterochromatin-dependent epimutants resistant to caffeine arise in fission yeast grown with threshold levels of caffeine. Isolates with unstable resistance have distinct heterochromatin islands with reduced expression of embedded genes, including some whose mutation confers caffeine resistance. Forced heterochromatin formation at implicated loci confirms that resistance results from heterochromatin-mediated silencing. Our analyses reveal that epigenetic processes promote phenotypic plasticity, letting wild-type cells adapt to unfavourable environments without genetic alteration. In some isolates, subsequent or coincident gene-amplification events augment resistance. Caffeine affects two anti-silencing factors: Epe1 is downregulated, reducing its chromatin association, and a shortened isoform of Mst2 histone acetyltransferase is expressed. Thus, heterochromatin-dependent epimutation provides a bet-hedging strategy allowing cells to adapt transiently to insults while remaining genetically wild type. Isolates with unstable caffeine resistance show cross-resistance to antifungal agents, suggesting that related heterochromatin-dependent processes may contribute to resistance of plant and human fungal pathogens to such agents.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequence data generated in this study have been submitted to GEO under accession number GSE138436. Source data are provided with this paper.
Code availability
The complete Workflow Description Language (WDL) pipeline script used for ChIP–seq and variation analyses is available at https://github.com/SitoTorres/Torres-Garcia-et-al.-2019.
References
Bannister, A. J. et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124 (2001).
Lachner, M., O’Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116–120 (2001).
Allshire, R. C. & Madhani, H. D. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19, 229–244 (2018).
Audergon, P. N. C. B. et al. Epigenetics. Restricted epigenetic inheritance of H3K9 methylation. Science 348, 132–135 (2015).
Ragunathan, K., Jih, G. & Moazed, D. Epigenetics. Epigenetic inheritance uncoupled from sequence-specific recruitment. Science 348, 1258699 (2015).
Jeggo, P. A. & Holliday, R. Azacytidine-induced reactivation of a DNA repair gene in Chinese hamster ovary cells. Mol. Cell. Biol. 6, 2944–2949 (1986).
Oey, H. & Whitelaw, E. On the meaning of the word ‘epimutation’. Trends Genet. 30, 519–520 (2014).
Zhang, K., Mosch, K., Fischle, W. & Grewal, S. I. S. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat. Struct. Mol. Biol. 15, 381–388 (2008).
Zofall, M. et al. RNA elimination machinery targeting meiotic mRNAs promotes facultative heterochromatin formation. Science 335, 96–100 (2012).
Wang, J., Reddy, B. D. & Jia, S. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. eLife 4, e06179 (2015).
Parsa, J.-Y., Boudoukha, S., Burke, J., Homer, C. & Madhani, H. D. Polymerase pausing induced by sequence-specific RNA-binding protein drives heterochromatin assembly. Genes Dev. 32, 953–964 (2018).
Sorida, M. et al. Regulation of ectopic heterochromatin-mediated epigenetic diversification by the JmjC family protein Epe1. PLoS Genet. 15, e1008129 (2019).
Yamanaka, S. et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature 493, 557–560 (2013).
Gallagher, P. S. et al. Iron homeostasis regulates facultative heterochromatin assembly in adaptive genome control. Nat. Struct. Mol. Biol. 25, 372–383 (2018).
Calvo, I. A. et al. Genome-wide screen of genes required for caffeine tolerance in fission yeast. PLoS ONE 4, e6619 (2009).
Ivanova, A. V., Bonaduce, M. J., Ivanov, S. V. & Klar, A. J. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 19, 192–195 (1998).
Nakayama, J., Rice, J. C., Strahl, B. D., Allis, C. D. & Grewal, S. I. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292, 110–113 (2001).
Kudo, N., Taoka, H., Toda, T., Yoshida, M. & Horinouchi, S. A novel nuclear export signal sensitive to oxidative stress in the fission yeast transcription factor Pap1. J. Biol. Chem. 274, 15151–15158 (1999).
Castillo, E. A., Vivancos, A. P., Jones, N., Ayté, J. & Hidalgo, E. Schizosaccharomyces pombe cells lacking the Ran-binding protein Hba1 show a multidrug resistance phenotype due to constitutive nuclear accumulation of Pap1. J. Biol. Chem. 278, 40565–40572 (2003).
Zofall, M., Smith, D. R., Mizuguchi, T., Dhakshnamoorthy, J. & Grewal, S. I. S. Taz1-Shelterin promotes facultative heterochromatin assembly at chromosome-internal sites containing late replication origins. Mol. Cell 62, 862–874 (2016).
Angerer, H. Eukaryotic LYR proteins interact with mitochondrial protein complexes. Biology (Basel) 4, 133–150 (2015).
Wang, S. W., Norbury, C., Harris, A. L. & Toda, T. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 112, 927–937 (1999).
Libuda, D. E. & Winston, F. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443, 1003–1007 (2006).
Møller, H. D., Parsons, L., Jørgensen, T. S., Botstein, D. & Regenberg, B. Extrachromosomal circular DNA is common in yeast. Proc. Natl Acad. Sci. USA 112, E3114–E3122 (2015).
Hull, R. M. et al. Transcription-induced formation of extrachromosomal DNA during yeast ageing. PLoS Biol. 17, e3000471 (2019).
Wu, S. et al. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature 575, 699–703 (2019).
Stajic, D., Perfeito, L. & Jansen, L. E. T. Epigenetic gene silencing alters the mechanisms and rate of evolutionary adaptation. Nat. Ecol. Evol. 3, 491–498 (2019).
Thodberg, M. et al. Comprehensive profiling of the fission yeast transcription start site activity during stress and media response. Nucleic Acids Res. 47, 1671–1691 (2019).
Yan, Y., Barlev, N. A., Haley, R. H., Berger, S. L. & Marmorstein, R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol. Cell 6, 1195–1205 (2000).
Cubas, P., Vincent, C. & Coen, E. An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161 (1999).
Heard, E. & Martienssen, R. A. Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157, 95–109 (2014).
Calo, S. et al. Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513, 555–558 (2014).
Antequera, F., Tamame, M., Villanueva, J. R. & Santos, T. DNA methylation in the fungi. J. Biol. Chem. 259, 8033–8036 (1984).
Wilkinson, C. R., Bartlett, R., Nurse, P. & Bird, A. P. The fission yeast gene pmt1 + encodes a DNA methyltransferase homologue. Nucleic Acids Res. 23, 203–210 (1995).
Fisher, M. C., Hawkins, N. J., Sanglard, D. & Gurr, S. J. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742 (2018).
Stone, N. R. et al. Dynamic ploidy changes drive fluconazole resistance in human cryptococcal meningitis. J. Clin. Invest. 129, 999–1014 (2019).
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 (1991).
Harigaya, Y. et al. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 442, 45–50 (2006).
Watson, A. T. et al. Optimisation of the Schizosaccharomyces pombe urg1 expression system. PLoS ONE 8, e83800 (2013).
Delerue, T. et al. Loss of Msp1p in Schizosaccharomyces pombe induces a ROS-dependent nuclear mutator phenotype that affects mitochondrial fission genes. FEBS Lett. 590, 3544–3558 (2016).
Bähler, J. et al. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951 (1998).
Tong, P. et al. Interspecies conservation of organisation and function between nonhomologous regional centromeres. Nat. Commun. 10, 2343 (2019).
Nerusheva, O. O., Galander, S., Fernius, J., Kelly, D. & Marston, A. L. Tension-dependent removal of pericentromeric shugoshin is an indicator of sister chromosome biorientation. Genes Dev. 28, 1291–1309 (2014).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–W165 (2016).
Diaz, A., Park, K., Lim, D. A. & Song, J. S. Normalization, bias correction, and peak calling for ChIP-seq. Stat. Appl. Genet. Mol. Biol. 11, 9 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Phanstiel, D. H., Boyle, A. P., Araya, C. L. & Snyder, M. P. Sushi.R: flexible, quantitative and integrative genomic visualizations for publication-quality multi-panel figures. Bioinformatics 30, 2808–2810 (2014).
Jeffares, D. C. et al. The genomic and phenotypic diversity of Schizosaccharomyces pombe. Nat. Genet. 47, 235–241 (2015).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 43, 11.10.1–11.10.33 (2013).
McLaren, W. et al. The Ensembl Variant Effect Predictor. Genome Biol. 17, 122 (2016).
Talevich, E., Shain, A. H., Botton, T. & Bastian, B. C. CNVkit: genome-wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput. Biol. 12, e1004873 (2016).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLOS Comput. Biol. 9, e1003118 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhu, A., Ibrahim, J. G. & Love, M. I. Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092 (2019).
Fletcher, S. J., Boden, M., Mitter, N. & Carroll, B. J. SCRAM: a pipeline for fast index-free small RNA read alignment and visualization. Bioinformatics 34, 2670–2672 (2018).
Braun, S. et al. The Cul4-Ddb1(Cdt)2 ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144, 41–54 (2011).
Woods, A. et al. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93, 491–500 (1989).
Pidoux, A. L. & Armstrong, J. The BiP protein and the endoplasmic reticulum of Schizosaccharomyces pombe: fate of the nuclear envelope during cell division. J. Cell Sci. 105, 1115–1120 (1993).
Acknowledgements
We thank L. Di Pompeo, A. Fellas and R. Yeboah for laboratory support; P. Tong, M. Lafos, R. Ard and S. Webb (Wellcome Centre for Cell Biology Bioinformatics Core) for sharing technical expertise; D. Kelly (Wellcome Centre Optical Instrumentation Laboratory) for microscopy and instrumentation support; members of the Allshire laboratory for valuable discussions; A. Bird, W. Bickmore and L. Massari for comments on the manuscript; T. Urano for the 5.1.1 (H3K9me) antibody; Y. Watanabe for the pRAD21 plasmid; K. Gull for the α-tubulin antibody; A. Marston for the Sgo1–GFP S. cerevisiae strain, K. Sawin for the Cdc11 antibody; and Edinburgh Genomics (NERC, R8/H10/56; MRC, MR/K001744/1; BBSRC, BB/J004243/1) and the Genetics Core, Edinburgh Clinical Research Facility, University of Edinburgh, for sequencing. S.T.-G. is supported by the Darwin Trust of Edinburgh. P.N.C.B.A. was supported by the Wellcome 4 Year PhD program in Cell Biology (093852). I.Y. is supported by an EMBO Long Term Fellowship (EMBO ALTF 130-2018). R.C.A. is a Wellcome Principal Research Fellow (095021, 200885); the Wellcome Centre for Cell Biology is supported by core funding from Wellcome (203149).
Author information
Authors and Affiliations
Contributions
S.T.-G., P.N.C.B.A. and R.C.A. conceived the project. S.T.-G. and P.N.C.B.A. performed preliminary studies. S.T.-G. performed experiments and bioinformatics. M.S. designed cup1-3xDSR experiments and contributed to ChIP–seq and qChIP experiments. A.L.P. performed cytology, cup1-TT and eccDNA experiments. I.Y. constructed strains expressing epitope-tagged Epe1 and Mst2 and performed western analysis. S.A.W. generated strains expressing Cup1–GFP and mutant Cup1 and contributed to Epe1 and Mst2 experiments. S.T.-G., A.L.P. and R.C.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Songtao Jia and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Identification of heterochromatin-dependent epimutants resistant to caffeine.
a, Frequencies of unstable (UR) and stable (SR) caffeine-resistant isolates obtained from three independent screens. 64% of isolates did not display a clear phenotype (unclear). b, Unstable (UR) and stable (SR) caffeine-resistant isolates were identified using this screening strategy. After growth on non-selective media for 14 d, caffeine resistance is lost in UR isolates but not in SR isolates. c, Caffeine resistance is lost progressively in unstable (UR) isolates but maintained in stable (SR) isolates. d, Caffeine resistance in UR isolates depends on the Clr4 H3K9 methyltransferase. clr4+ (clr4Δ) or an unlinked intergenic region (controlΔ) were deleted in unstable (UR-2) and stable (SR-2) caffeine-resistant isolates. e, A mutation in pap1+ confers caffeine resistance in the stable isolate SR-1. Left, whole-genome sequencing of the stable isolate SR-1 revealed a 7-nucleotide insertion in pap1+. The insertion results in a truncated Pap1 protein (Pap1-N424STOP) that lacks the nuclear export signal (NES). CRD, cysteine-rich domain. Right, Pap1-N424STOP is resistant to caffeine. The 7-nucleotide insertion identified in SR-1 was introduced into the pap1+ gene of wild-type cells (Pap1-N424STOP) and caffeine resistance assessed. hba1Δ and SR-1 cells were used as positive controls. Experiments in b–d and e, right, were independently repeated at least twice with similar results.
Extended Data Fig. 2 Unstable (UR) caffeine-resistant isolates are bona fide epimutants.
a–e, Genetic changes (clr5-Q264STOP meu27-S100Y) found in 4 of 30 unstable isolates do not contribute to the caffeine-resistant phenotype or cause the formation of ectopic heterochromatin. a, Whole-genome sequencing of unstable isolates UR-1, UR-3, UR-5 and UR-7 revealed single-nucleotide polymorphisms (SNPs) in clr5+ (clr5-Q264STOP) and in meu27+ (meu27-S100Y). b, Left, schematic of experiment to determine whether clr5-Q264STOP meu27-S100Y cells form more caffeine-resistant colonies than wild-type cells. Wild-type (wt) and clr5-Q264STOP meu27-S100Y cells were plated on +CAF medium (105 cells per plate, 20 plates per strain). Caffeine-resistant colonies were counted after 7 d. Right, clr5-Q264STOP meu27-S100Y form a similar number of caffeine-resistant colonies to wt cells. Data are mean of 20 technical replicates. P value from a two-tailed Student’s t-test is indicated. c, clr5-Q264STOP meu27-S100Y cells are not resistant to caffeine. clr5-Q264STOP meu27-S100Y cells were serially diluted and spotted on –CAF and +CAF plates to assess caffeine resistance. hba1Δ cells served as a positive control. Experiment was independently repeated at least twice with similar results. d, Genome-wide H3K9me2 ChIP-seq enrichment in wt and clr5-Q264STOP meu27-S100Y cells. Data are represented as relative fold enrichment over input. e, H3K9me2 ChIP-seq enrichment at known heterochromatin islands detected in epe1Δ cells9 in wt and clr5-Q264STOP meu27-S100Y cells. Data are represented as relative fold enrichment over input. f, Gene transcript levels within and flanking ectopic heterochromatin islands in individual isolates. See Fig. 2b. Data are mean ± s.d. from three biological replicates. P values <0.05 from a two-tailed Student’s t-test are indicated.
Extended Data Fig. 3 Twenty-four of 30 unstable (UR) caffeine-resistant isolates display an ectopic heterochromatin island over the ncRNA.394 locus.
a, H3K9me2 ChIP-seq enrichment at the ncRNA.394 locus in individual isolates (left, coverage tracks; right, heatmaps). Data are represented as relative fold enrichment over input. Relevant genes within and flanking ectopic heterochromatin islands are indicated. Red arrows indicate essential genes. Dumbbells indicate primer pairs used in b, c and e. b, Quantitative chromatin immunoprecipitation (qChIP) of H3K9me2 levels on ncRNA.394 in individual isolates. Primer pairs used are indicated in a (ncRNA.394, primer pair 5). c, SPBC17G9.13c+ gene transcript levels in individual isolates. P values from a two-tailed Student’s t-test are indicated. Primer pairs used are indicated in a (SPBC17G9.13c+, primer pair 3). d, Deletion of ncRNA.394 or non-essential adjacent genes does not result in caffeine resistance. Experiment was independently repeated at least twice with similar results. e, qChIP of H3K9me2 levels at the ncRNA.394 locus in UR-2 cells. UR-2 cells were grown in the absence (–CAF) or presence (+CAF) of caffeine overnight or in the absence of caffeine for 14 d (+14day/–CAF). Primer pairs used are indicated in a. In b, c, e, data are mean ± s.d. from three biological replicates.
Extended Data Fig. 4 Forced synthetic heterochromatin targeting to the identified loci is sufficient to drive caffeine resistance in wild-type cells.
a–c, Quantitative chromatin immunoprecipitation (qChIP) of H3K9me2 levels in wild-type (wt) cells harbouring 4xtetO binding sites at the identified ectopic heterochromatin loci (or ura4 as control) and expressing TetR–Clr4* in the absence or presence of AHT. a, hba1 locus. b, ncRNA.394 locus. c, ura4 locus. Data are mean ± s.d. from three biological replicates. Dumbbells indicate primer pairs used. Red arrows indicate essential genes. d, Forced synthetic heterochromatin targeting to the mbx2 locus is sufficient to drive caffeine resistance in wt cells. qChIP of H3K9me2 levels in wt cells harbouring 4xtetO binding sites at the mbx2 ectopic heterochromatin locus and expressing TetR–Clr4* in the absence or presence of AHT. Data are mean ± s.d. from three biological replicates. Dumbbells indicate primer pairs used. e, Strains from a–c were assessed for resistance to the antifungal agents tebuconazole (+TEZ) and fluconazole (+FLZ). Experiments were independently repeated at least twice with similar results.
Extended Data Fig. 5 Unstable (UR) caffeine-resistant isolates show cross-resistance to antifungals and siRNA generation at ectopic heterochromatin islands.
a, Unstable caffeine-resistant isolates UR-1 and UR-2 were serially diluted and spotted on non-selective (N/S), caffeine (+CAF), clotrimazole (+CLZ), tebuconazole (+TEZ) and fluconazole (+FLZ) medium to assess resistance. Experiment was independently repeated at least twice with similar results. b, c, Left, small RNA sequencing detects siRNAs (21–24 nucleotides) homologous to ectopic heterochromatin islands in UR-1 (b, hba1 locus) and UR-2 (c, ncRNA.394 locus) compared to wild-type (wt) cells. Right, siRNAs mapping to pericentromeric dgI/dhI repeats of chromosome I shown as control. Sequencing was performed once. *, transcripts mapping to the highly expressed gene eno101+ in euchromatic wild-type conditions (note these are unidirectional RNAs and not siRNAs). d, Caffeine resistance depends on RNAi. dcr1+ (dcr1Δ), ago1+ (ago1Δ) or an unlinked intergenic region (controlΔ) were deleted in UR-2 cells. Experiments were independently repeated at least twice with similar results.
Extended Data Fig. 6 Decreased cup1+ transcript levels or Cup1 LYR-domain mutation results in caffeine resistance.
a, An additional copy of cup1+ with 3× determinant of selective removal (DSR) motifs fused to its 3′ untranslated region was inserted at an intergenic region (LocusPX:cup1-3xDSR). Bottom left, after deletion of endogenous cup1+, cells expressing only cup1-3xDSR were assessed for caffeine resistance. Bottom right, transcript levels of cup1+ and SPBC17G9.12c+ (as control) in cup1Δ locusPX:cup1-3xDSR cells compared to wild-type. Data are mean ± s.d. from three biological replicates. P value from a two-tailed Student’s t-test is indicated. Dumbbells indicate primer pairs used. b, The 144-bp transcriptional terminator site from ura4+ was inserted in place of part of the putative cup1+ promoter (cup1-TT). Bottom left, cells were assessed for caffeine resistance. Bottom right, transcript levels of cup1+ and SPBC17G9.12c+ (as control) in cup1-TT cells compared to wild-type. Data are mean ± s.d. from three biological replicates. P value from a two-tailed Student’s t-test is indicated. Dumbbells indicate primer pairs used. c, Cup1 localizes to mitochondria. Cells expressing either untagged Cup1 (top row) or Cup1–GFP (bottom three rows) were fixed and processed for immunofluorescence with anti-GFP antibody and Alexa-488 secondary antibody and DNA was stained with DAPI. The mitochondrial protein Arg11–mCherry served as a positive control for mitochondrial localization. All images in the green channel (Cup1–GFP) are scaled relative to each other, as are those in the red channel (Arg11–mCherry); DAPI images are autoscaled. Bar, 5 μm. d, Point mutations (L73G and F99G) were introduced in the LYR domain of Cup1 and cells were assessed for caffeine resistance. Mutations were designed based on Phyre2 tool analysis. hba1Δ cells were used as positive control. Experiments in c and d were independently repeated at least twice with similar results.
Extended Data Fig. 7 CNV analysis reveals a partial duplication of chromosome III in 12 of 30 unstable (UR) caffeine-resistant isolates.
a, Chromosome III coverage plots with overlaid segments in UR isolates showing partial duplication of chromosome III. Location of cds1+ is highlighted. Wild-type ChIP-seq input data were used as the reference. b–d, Epigenetic changes preceded genetic changes (CNV) in unstable caffeine-resistant isolate UR-2. b, H3K9me2 ChIP-seq enrichment at the ncRNA.394/cup1 locus (left) and chromosome III coverage plots with overlaid segments (right) in UR-2 (4day/+CAF) cells and following their prolonged growth on +CAF for an additional 3 d (7day/+CAF). Wild-type ChIP-seq input data were used as the reference for CNV analysis. c, clr4+ (clr4Δ) or an unlinked intergenic region (controlΔ) were deleted in UR-2 cells (4day/+CAF) and UR-2 (7day/+CAF). All (6/6) UR-2 (4day/+CAF) clr4Δ transformants lost resistance to caffeine, whereas only 50% (3/6, transformants 1, 4 and 5) UR-2 (7day/+CAF) lost resistance to caffeine. Experiments were independently repeated at least twice with similar results. cds1+ DNA levels in extracted genomic DNA were assessed by qPCR. Data are mean ± s.d. from three biological replicates. d, H3K9me2 ChIP-seq enrichment at the ncRNA.394/cup1 locus (left) and chromosome III coverage plots with overlaid segments (right) in UR-2 (7day/+CAF) cells and following their prolonged growth on non-selective medium for 14 days (7day/+CAF→14day/–CAF). Wild-type ChIP-seq input data were used as the reference for CNV analysis.
Extended Data Fig. 8 CNV of chromosome III corresponds to extrachromosomal circular DNA (eccDNA).
a, b, Junctions of putative extrachromosomal circles were identified at repetitive sequences by inspection of CNV plots for UR-2 (7day/+CAF) (a) and UR-4 (b). In maps and lower panels, positions of 5S rRNA.24 and 5S rRNA.26 (pink arrows), LTR3 and LTR27 (green arrows) and flanking genes are indicated. PCR primers (half arrows) flanking 5S rRNA.24 (A (forward); B1,2 (reverse)) and 5S rRNA.26 (C1,2; D1,2) were used to amplify products from wild-type (wt) and UR-2 (7day/+CAF) ChIP input samples, along with primer combinations (C1,2; B1,2) specific for the putative circle junctions (vertical black lines). Primers flanking LTR3 (E; F1,2) and LTR27 (G1,2; H) were used to amplify products from wild-type and UR-4 ChIP input samples, along with primer combinations (G1,2; F1,2) specific for the putative circle junction. Shaded boxes indicate primer locations and predicted circle junctions (pink: 5S rRNA.24/26, green: LTR3/27). Right, restriction enzyme-digested genomic DNA isolated from wild-type (wt), UR-2 (7day/+CAF) and UR-4 was separated on an ethidium bromide (EtBr)-containing gel followed by Southern analysis using the indicated probes (925, blue; 520, purple; 44, red). Relevant restriction enzyme sites are indicated. Predicted sizes of hybridizing fragments and DNA size markers are indicated (kb). PCR experiments were independently repeated at least twice with similar results. For gel source data, see Supplementary Fig. 1b.
Extended Data Fig. 9 The heterochromatin profile of low-caffeine-treated wild-type cells resembles that of untreated epe1Δ cells.
a, Growth of cells in caffeine. Wild-type (wt) cells were grown in the presence of low (7 mM) or medium (14 mM) caffeine for 18 h. Cell number was counted every 6 h. Note: a larger inoculum was used for 14 mM caffeine culture to obtain an equivalent final number of cells. Data are mean ± s.d. from three biological replicates. Cells from the 18-h time point were used for d. b, c, H3K9me2 ChIP-seq enrichment at previously-detected facultative heterochromatin loci (described in ref. 9 (b and c), ref. 13 (b), ref. 10 (b), ref. 12 (b) and ref. 14 (b)), in wt cells treated with low or medium dose of caffeine or low dose (1 mM) of H2O2, compared to untreated epe1Δ and wt cells. Data are represented as relative fold enrichment over input. A subset of facultative heterochromatin loci detected in untreated epe1Δ cells (refs.9,10,12) was detected in low-caffeine-treated wt cells. Asterisks in c indicate loci with similar H3K9me2 patterns in low-caffeine-treated wt cells and untreated epe1Δ cells, but not untreated wt cells. Facultative heterochromatin loci formed in the absence of the exosome (ref.13) or in wt cells grown at 18 °C (ref. 14) were not detected in wt cells treated with low or medium caffeine or low H2O2. d, Quantitative ChIP (qChIP) of H3K9me2 levels on ncRNA.394/cup1 in wt cells following 18 h exposure to low or medium caffeine. H3K9me2 levels were normalized to S. octosporus spike-in control. Data are mean ± s.d. from three biological replicates. e, H3K9me2 ChIP-seq enrichment at ncRNA.394/cup1 and mcp7 loci (or at pericentromeric dgI/dhI repeats of chromosome I as control) in wt cells following 18 h exposure to low H2O2. Data are represented as relative fold enrichment over input. Red arrows indicate essential genes. Lower levels of H3K9me2 at pericentromeric repeats upon H2O2 treatment may be due to H2O2-specific regulation of limiting heterochromatin factors at this locus. f, epe1+ RNA levels do not change upon caffeine treatment. Total RNA-seq of wt cells treated with low caffeine. Transcripts encoding components of the Clr4 H3K9 methyltransferase CLRC complex (clr4+, rik1+, raf1+, raf2+, pcu4+ and rbx1+) and the antisilencing factors epe1+ and mst2+ are highlighted. Experiment was independently repeated twice with similar results. g, epe1Δ cells display increased resistance to caffeine. Left, schematic of experiment. Wild-type, epe1Δ and clr4Δ cells were plated on +CAF medium (105 cells/plate, 40 plates/strain). Caffeine-resistant colonies were counted after 7 d. Right, compared to wt cells, epe1Δ forms more, whereas clr4Δ forms fewer, caffeine-resistant colonies. Note that the total number of resistant colonies also includes genetic mutants. Data are mean from forty technical replicates. P values from a two-tailed Student’s t-test are indicated.
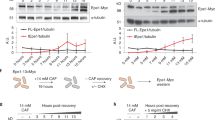
Extended Data Fig. 10 A shortened version of the anti-silencing factor Mst2 is produced upon exposure to caffeine.
a, Western analysis of Mst2-13xMyc (left) and Gcn5-13xMyc (as HAT control, right) before and after caffeine treatment (medium concentration, 14 mM). Tagged proteins are expressed from their endogenous loci. Loading controls: left, Bip1; right, Cdc11. Experiments were independently repeated at least twice with similar results. For gel source data, see Supplementary Fig. 1c. b, Total RNA-seq for mst2 (left) and gcn5 (as HAT control, right) of untreated wild-type cells (top) or wild-type cells treated with medium caffeine concentration (bottom). Diagrams illustrate mst2 and gcn5 transcripts and predicted protein domains. Reads are normalized to RPKM. Red dashed lines indicate the region of full length mst2 transcript absent from the short isoform. The MYST zinc finger (ZnF) domain, required for S. cerevisiae Esa1 acetyltransferase activity29, is truncated in the short isoform of Mst2. The alternative mst2 TSS used in caffeine conditions was previously annotated28. Experiment was independently repeated twice with similar results.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1. a, Source gel data for Fig. 4c. b, Source gel data for Extended Data Fig. 8. c, Source gel data for Extended Data Fig. 10a. Yellow and dashed boxes indicate data used in figures. Supplementary Table 1. Summary of epigenetic (H3K9me2 domains) and genetic (SNPs, indels and copy number variation) changes found in unstable (UR) caffeine-resistant isolates. Supplementary Table 2. Schizosaccharomyces pombe strains used in this study. Supplementary Table 3. Oligonucleotides used in this study. Supplementary Table 4. Genomic coordinates used to generate heatmaps for heterochromatin islands.
Rights and permissions
About this article
Cite this article
Torres-Garcia, S., Yaseen, I., Shukla, M. et al. Epigenetic gene silencing by heterochromatin primes fungal resistance. Nature 585, 453–458 (2020). https://doi.org/10.1038/s41586-020-2706-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2706-x
This article is cited by
-
Evolution beyond DNA: epigenetic drivers for evolutionary change?
BMC Biology (2023)
-
DNA methylation regulator-mediated modification patterns and risk of intracranial aneurysm: a multi-omics and epigenome-wide association study integrating machine learning, Mendelian randomization, eQTL and mQTL data
Journal of Translational Medicine (2023)
-
The phenomenon of strain degeneration in biotechnologically relevant fungi
Applied Microbiology and Biotechnology (2023)
-
Molecular mechanisms of transgenerational epigenetic inheritance
Nature Reviews Genetics (2022)
-
Proteasome-dependent truncation of the negative heterochromatin regulator Epe1 mediates antifungal resistance
Nature Structural & Molecular Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.