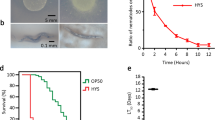
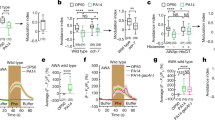
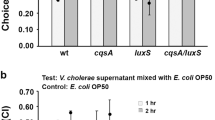
Abstract
Caenorhabditis elegans must distinguish pathogens from nutritious food sources among the many bacteria to which it is exposed in its environment1. Here we show that a single exposure to purified small RNAs isolated from pathogenic Pseudomonas aeruginosa (PA14) is sufficient to induce pathogen avoidance in the treated worms and in four subsequent generations of progeny. The RNA interference (RNAi) and PIWI-interacting RNA (piRNA) pathways, the germline and the ASI neuron are all required for avoidance behaviour induced by bacterial small RNAs, and for the transgenerational inheritance of this behaviour. A single P. aeruginosa non-coding RNA, P11, is both necessary and sufficient to convey learned avoidance of PA14, and its C. elegans target, maco-1, is required for avoidance. Our results suggest that this non-coding-RNA-dependent mechanism evolved to survey the microbial environment of the worm, use this information to make appropriate behavioural decisions and pass this information on to its progeny.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data are available from: BioProject under accession number PRJNA553700. Any data related to the study that are not provided in the Article and its Supplementary Information can be obtained upon reasonable request from the corresponding author.
References
Samuel, B. S., Rowedder, H., Braendle, C., Félix, M.-A. & Ruvkun, G. Caenorhabditis elegans responses to bacteria from its natural habitats. Proc. Natl Acad. Sci. USA 113, E3941–E3949 (2016).
Zhang, Y., Lu, H. & Bargmann, C. I. Pathogenic bacteria induce aversive olfactory learning in Caenorhabditis elegans. Nature 438, 179–184 (2005).
Moore, R. S., Kaletsky, R. & Murphy, C. T. Piwi/PRG-1 Argonaute and TGF-β mediate transgenerational learned pathogenic avoidance. Cell 177, 1827–1841.e12 (2019).
Meisel, J. D., Panda, O., Mahanti, P., Schroeder, F. C. & Kim, D. H. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell 159, 267–280 (2014).
Kim, D. H. et al. A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297, 623–626 (2002).
Melo, J. A. & Ruvkun, G. Inactivation of conserved C. elegans genes engages pathogen- and xenobiotic-associated defenses. Cell 149, 452–466 (2012).
Lee, K. & Mylonakis, E. An intestine-derived neuropeptide controls avoidance behavior in Caenorhabditis elegans. Cell Rep. 20, 2501–2512 (2017).
Estes, K. A., Dunbar, T. L., Powell, J. R., Ausubel, F. M. & Troemel, E. R. bZIP transcription factor zip-2 mediates an early response to Pseudomonas aeruginosa infection in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA 107, 2153–2158 (2010).
Troemel, E. R. et al. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet. 2, e183 (2006).
Ghildiyal, M. & Zamore, P. D. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108 (2009).
Winston, W. M., Sutherlin, M., Wright, A. J., Feinberg, E. H. & Hunter, C. P. Caenorhabditis elegans SID-2 is required for environmental RNA interference. Proc. Natl Acad. Sci. USA 104, 10565–10570 (2007).
Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366 (2001).
Ketting, R. F. et al. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659 (2001).
McEwan, D. L., Weisman, A. S. & Hunter, C. P. Uptake of extracellular double-stranded RNA by SID-2. Mol. Cell 47, 746–754 (2012).
Kaletsky, R. et al. Transcriptome analysis of adult Caenorhabditis elegans cells reveals tissue-specific gene and isoform expression. PLoS Genet. 14, e1007559 (2018).
Winston, W. M., Molodowitch, C. & Hunter, C. P. Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 (2002).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Tops, B. B. J. et al. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 33, 347–355 (2005).
Tabara, H., Yigit, E., Siomi, H. & Mello, C. C. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell 109, 861–871 (2002).
Ketting, R. F., Haverkamp, T. H. A., van Luenen, H. G. A. M. & Plasterk, R. H. A. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99, 133–141 (1999).
Posner, R. et al. Neuronal small RNAs control behavior transgenerationally. Cell 177, 1814–1826.e15 (2019).
Liu, H. et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 3, 1073 (2012).
Grishok, A. et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34 (2001).
Sowa, J. N. et al. The Caenorhabditis elegans RIG-I homolog DRH-1 mediates the intracellular pathogen response upon viral infection. J. Virol. 94, e01173-19 (2020).
Ashe, A. et al. A deletion polymorphism in the Caenorhabditis elegans RIG-I homolog disables viral RNA dicing and antiviral immunity. eLife 2, e00994 (2013).
Welker, N. C. et al. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 16, 893–903 (2010).
Aoki, K., Moriguchi, H., Yoshioka, T., Okawa, K. & Tabara, H. In vitro analyses of the production and activity of secondary small interfering RNAs in C. elegans. EMBO J. 26, 5007–5019 (2007).
Smardon, A. et al. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178 (2000).
Couteau, F., Guerry, F., Müller, F. & Palladino, F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 3, 235–241 (2002).
Austin, J. & Kimble, J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 51, 589–599 (1987).
Ouyang, J. P. T. et al. P granules protect RNA interference genes from silencing by piRNAs. Dev. Cell 50, 716–728.e6 (2019).
Wurtzel, O. et al. The single-nucleotide resolution transcriptome of Pseudomonas aeruginosa grown in body temperature. PLoS Pathog. 8, e1002945 (2012).
Livny, J., Brencic, A., Lory, S. & Waldor, M. K. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 34, 3484–3493 (2006).
Zhan, Y. et al. NfiR, a new regulatory noncoding RNA (ncRNA), is required in concert with the NfiS ncRNA for optimal expression of nitrogenase genes in Pseudomonas stutzeri A1501. Appl. Environ. Microbiol. 85, e00762-19 (2019).
Kuvbachieva, A. et al. Identification of a novel brain-specific and Reelin-regulated gene that encodes a protein colocalized with synapsin. Eur. J. Neurosci. 20, 603–610 (2004).
Miyara, A. et al. Novel and conserved protein macoilin is required for diverse neuronal functions in Caenorhabditis elegans. PLoS Genet. 7, e1001384 (2011).
Arellano-Carbajal, F. et al. Macoilin, a conserved nervous system-specific ER membrane protein that regulates neuronal excitability. PLoS Genet. 7, e1001341 (2011).
Neal, S. J. et al. A forward genetic screen for molecules involved in pheromone-induced dauer formation in Caenorhabditis elegans. G3 6, 1475–1487 (2016).
Hudzik, C., Hou, Y., Ma, W. & Axtell, M. J. Exchange of small regulatory RNAs between plants and their pests. Plant Physiol. 182, 51–62 (2020).
Weiberg, A. et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 (2013).
Palominos, M. F. et al. Transgenerational diapause as an avoidance strategy against bacterial pathogens in Caenorhabditis elegans. MBio 8, e01234-17 (2017).
Burton, N. O. et al. Cysteine synthases CYSL-1 and CYSL-2 mediate C. elegans heritable adaptation to P. vranovensis infection. Nat. Commun. 11, 1741 (2020).
Hmelo, L. R. et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protocols 10, 1820–1841 (2015).
Chambers, J. R. & Sauer, K. The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ. J. Bacteriol. 195, 4678–4688 (2013).
Guzman, L. M., Belin, D., Carson, M. J. & Beckwith, J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 (1995).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Ho, J., Tumkaya, T., Aryal, S., Choi, H. & Claridge-Chang, A. Moving beyond P values: data analysis with estimation graphics. Nat. Methods 16, 565–566 (2019).
Acknowledgements
We thank the C. elegans Genetics Center for strains; the Genomics Core Facility at Princeton University; BioRender for model figure design software; W. Wang for helping to develop methods to sequence bacterial small RNAs; the laboratory of C.T.M. for discussion; and R. Clausen and J. Ashraf for assistance. C.T.M. is the Director of the Glenn Center for Aging Research at Princeton and an HHMI-Simons Faculty Scholar. This work was supported by a Pioneer Award to C.T.M. (NIGMS DP1GM119167), the Glenn Foundation for Medical Research (GMFR CNV1001899), the HHMI-Simons Faculty Scholar Program (AWD1005048), T32GM007388 (NIGMS) support of R.S.M. and G.D.V., and a Pioneer Award to Z.G. (DP1A1124669).
Author information
Authors and Affiliations
Contributions
R.K., R.S.M., G.D.V., Z.G. and C.T.M. designed experiments. R.K. and R.S.M. performed experiments and analysed data. G.D.V. constructed P11 E. coli and PA14 mutant strains. L.R.P. and R.K. analysed small-RNA-seq data. R.K., R.S.M. and C.T.M. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 PA14 sRNAs induce maternal PA14 avoidance and increased daf-7 expression in the ASI neurons.
a, Worms exposed to a PA14 bacterial lawn for 24 h learn to avoid PA14 in a choice assay, but PA14 DNA exposure alone does not induce maternal avoidance of PA14. b, Training with large RNA (>200 nt) isolated from bacterial lawns of PA14 is not sufficient for maternal PA14 avoidance. c, Bioanalyzer results of isolated PA14 total RNA and fractionated small and large RNAs. RNA levels were normalized for worm training. d, ΔlasR sRNA exposure does not induce PA14 learned avoidance. e, Development of progeny of PA14 sRNA-trained mothers was not delayed compared to progeny of OP50-trained mothers. n = 6 plates per condition with 23–142 progeny per plate, mean ± s.e.m. f, pmk-1(km25) worms learn to avoid PA14 when exposed to PA14 bacteria lawns. g, pmk-1(km25) is not required for PA14 sRNA-induced pathogenic learning. h, irg-1p::gfp expression is induced by PA14 bacterial lawn exposure, but not by PA14 sRNAs alone. i, GFP intensity from h was quantified, n = 33, 24, 43 and 35 worms, left to right. j, Model of PA14 bacteria lawn and sRNA learning. k, Location of ASI and ASJ neurons. l, The 24-h exposure of worms to a PA14 lawn or PA14 sRNA increases daf-7p::gfp expression in the ASI neurons. Two-way ANOVA, Tukey’s multiple comparison test. n = 35, 41, 41 and 49 neurons, left to right. m, ΔlasR sRNA exposure does not induce changes in daf-7p::gfp ASI expression. One-way ANOVA, Tukey’s multiple comparison test. n = 38, 27 and 29 neurons, left to right. Biological replicates: 2 (a), 3(b–d, f–i, m), 6 (l). For choice assays, each dot represents an individual choice assay plate (average of 115 worms per plate) with data shown from all replicates. For box plots, centre line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. One-way (d, m) and Two-way ANOVA (a, b, e–g, i, l), Tukey’s multiple comparison test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001, n.s., not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplementary Table 4 for exact sample sizes (n) and P values.
Extended Data Fig 2 sRNA and bacterial lawn training of C. elegans RNAi pathway mutants.
a, Wild-type and sid-2(qt42) worms were trained on OP50 or PA14 bacterial lawns for 24 h and tested for learned PA14 avoidance. b, c, Wild-type and dcr-1(mg375) worms were trained on OP50 or PA14 lawns (b) or OP50 or PA14 sRNA (c). d, Wild-type, dcr-1(mg375) and dcr-1(mg375);vha-6p::dcr-1 worms were trained on OP50 or PA14 bacterial lawns for 24 h and tested for learned PA14 avoidance. Training and choice assays for dcr-1(mg375) and dcr-1(mg375);vha-6p::dcr-1 worms were performed on the same plates. For choice assays, transgenic worms expressing the rescue construct and pharyngeal mCherry were counted using a fluorescence dissecting microscope. Non-transgenic, non-fluorescent dcr-1(mg375) sibling worms from the same plates were also counted and the results are shown. e, Wild-type and sid-1(pk3321) worms were trained on OP50 or PA14 bacterial lawns for 24 h and tested for learned PA14 avoidance. sid-1 mutants have a constitutively high naive avoidance, but are able to learn avoidance after training on a PA14 lawn, but not after training on sRNAs. f–j, Wild-type, rde-1(ne219) (f, g), rde-2(pk1657) (h, i) or rde-4(ne305) (j) worms were trained on OP50 or PA14 bacterial lawns (f, h, j) or sRNA (g, i) for 24 h and tested for learned PA14 avoidance. rde-1, rde-2 and rde-4 mutants are able to learn avoidance following bacteria lawn training, but do not additionally avoid PA14 following sRNA training. k, l, Wild-type and mut-7(pk720) worms were trained on OP50 or PA14 lawns (k) or sRNA (l) for 24 h and tested for learned PA14 avoidance. These mutants (in e–l) have high naive preference after training on sRNAs only. m–p, Wild-type and alg-1(gk214) (m, n) or drh-1(ok3495) (o, p) worms were trained on OP50 or PA14 bacterial lawns (m, o) or sRNA (n, p) for 24 h and tested for learned PA14 avoidance. Biological replicates: 3 (d, f, h–p), 4 (a–c, g), 6 (e). For choice assays, each dot represents an individual choice assay plate (average of 115 worms per plate) with data shown from all replicates. For box plots, centre line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-way ANOVA (a–p), Tukey’s multiple comparison test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001, n.s., not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplementary Table 4 for exact sample sizes (n) and P values.
Extended Data Fig. 3 sRNA and bacterial lawn training of C. elegans sRNA pathway and germline mutants.
a–e, Wild-type and prg-1(n4357) (a), prg-1 germline rescue (b), rrf-1(pk1417) (c), rrf-3(pk1426) (d), or hpl-2(ok916) worms (e) were trained on OP50 or PA14 bacterial lawns or sRNA (a, right) for 24 h and tested for learned PA14 avoidance. f, g, Representative images of OP50- or PA14-trained prg-1;daf-7::gfp (f) or glp-1;daf-7::gfp (g) mothers. ASI neuron = white arrow, ASJ neuron = grey arrow. h, Germline-less glp-1 mutants induce daf-7p::gfp expression in the ASJ neuron after PA14 lawn exposure. n = 42 (OP50), 46 (PA14) neurons, two-tailed Student’s t-test. i, Wild-type and meg-3(tm4259) meg-4(ax2026) worms were trained on OP50 or PA14 bacterial lawns for 24 h and tested for learned PA14 avoidance. Biological replicates: 3 (a–i). For choice assays, each dot represents an individual choice assay plate (average of 115 worms per plate) with data shown from all replicates. For box plots, centre line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-way ANOVA (a–e, i), Tukey’s multiple comparison test. ****P < 0.0001, n.s., not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplementary Table 4 for exact sample sizes (n) and P values.
Extended Data Fig. 4 sid-1, sid-2 and dcr-1 F1 progeny of plate and sRNA trained parent worms.
a–c, Progeny of sid-1 (a), sid-2 (b) and dcr-1 (c) mutants are defective in both transgenerational pathogen avoidance following maternal bacterial lawn (left) and PA14 sRNA training (right). Biological replicates: 3 (a–c). For choice assays, each dot represents an individual choice assay plate (average of 115 worms per plate) with data shown from all replicates. For box plots, centre line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-way ANOVA (a–c), Tukey’s multiple comparison test. **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001, n.s., not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplementary Table 4 for exact sample sizes (n) and P values.
Extended Data Fig. 5 RNA sequencing of sRNAs from PA14.
a, sRNA training protocol using RNA isolated from PA14 cultures grown at 25 °C or 15 °C on plates, or in liquid culture. b, Principal component analysis of PA14 sRNA sequencing. c, Upregulated sRNAs in 25-°C grown PA14 compared to 15 °C-grown and liquid-grown PA14 sRNAs. The outermost grey circle represents the PA14 genome, with previously identified sRNAs that were upregulated (DESeq2, adjusted P ≤ 0.05) in the 25-°C plate condition relative to 15-°C plate grown PA14 (pink) or liquid-grown PA14 (green) are indicated. Overlapping sRNAs upregulated in the 25-°C plate condition relative to both other conditions are noted (blue dots). sRNAs annotated with white lines. d, Venn diagram showing the number of PA14 sRNAs upregulated at 25-°C growth conditions compared to liquid growth (blue circle) or 15-°C growth (red circle). The six shared upregulated sRNAs are shown.
Extended Data Fig. 6 Identification and testing of the differentially regulated PA14 sRNA.
a, P11 expressed in E. coli induces PA14 avoidance in a choice assay between PA14 and E. coli strain MG1655. Worms were trained on OP50, PA14, E. coli strain MG1655 containing empty vector (control), or E. coli strain MG1655 expressing PA14 small RNAs (red). Following training, a choice assay was performed between E. coli strain MG1655 and PA14. Similar to Fig. 3c, worms trained on PA14 or E. coli expressing P11 exhibited PA14 avoidance behaviour. One-way ANOVA with Tukey’s multiple comparison test. b, PA14 plate-trained worms do not avoid E. coli expressing P11 compared to OP50 in a choice assay. Two-tailed Student’s t-test. c, PA14 ΔP11 bacteria grown on NGM plates produce pyocyanin (blue pigment on plates) similar to PA14. d, e, Model showing induced learning pathways for wild-type worms on isolated PA14 ΔP11 sRNA (d) or on PA14 ΔP11 bacteria lawns (e). f, Exposure to E. coli expressing P11 does not affect total brood size (left) (two-tailed Student’s t-test), or the number of progeny hatched per day (right). Two-way ANOVA with Tukey’s multiple comparison test. n = 15 worms were analysed per condition. g, Worms exposed to control (top) or E. coli expressing P11 appear healthy after 24 h of training. Biological replicates: 2 (a), 3 (b, c, g). For choice assays, each dot represents an individual choice assay plate (average of 115 worms per plate) with data shown from all replicates. For box plots, centre line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. **P ≤ 0.01, ****P < 0.0001, n.s., not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplementary Table 4 for exact sample sizes (n) and P values.
Extended Data Fig. 7 P11 region of homology with maco-1 and behaviour of vhp-1 mutants.
a, Wormbase depiction of the maco-1 genomic locus and the region of P11 homology (red). b, Model depicting induced learning pathways in wild-type or maco-1 mutant worms as a result of OP50 or PA14 lawn exposure. c, F1 progeny of OP50 or PA14 sRNA-exposed maco-1 mutant mothers were tested for PA14 avoidance behaviour. d, vhp-1expression is increased in PA14-exposed mothers, but not in F1 progeny3. Mean; DESeq2 adjusted P values are shown. P0, n = 6 replicates per condition; F1, n = 4 replicates per condition. e, The vhp-1(sa336) mutation is a null allele (early stop codon)40. Wild-type and vhp-1(sa366) worms were trained on OP50 or PA14 bacterial lawns for 24 h and tested for learned PA14 avoidance (top). f, Wild-type and vhp-1(sa366) worms were trained on OP50 or PA14 sRNA for 24 h and tested for learned PA14 avoidance. g, Progeny of vhp-1(sa366) PA14 lawn-trained worms inherit PA14 avoidance behaviour. h, F1 progeny of OP50 or PA14 lawn-exposed maco-1 mutant mothers were tested for PA14 avoidance behaviour. Biological replicates: 3 (c, e–h). For choice assays, each dot represents an individual choice assay plate (average of 115 worms per plate) with data shown from all replicates. For box plots, centre line is the median, box extends from the 25th to the 75th percentile; whiskers denote minimum–maximum values. Two-way ANOVA (c, e–h), Tukey’s multiple comparison test. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P < 0.0001, n.s., not significant. Mean differences are shown using Cumming estimation plots47, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. See Supplementary Table 4 for exact sample sizes (n) and P values.
Supplementary information
41586_2020_2699_MOESM1_ESM.pdf
Supplementary Information Estimation statistics for pooled and individual choice assay data. Mean differences are shown using Cumming estimation plots, with each graphed as a bootstrap sampling distribution. Mean differences are depicted as dots; 95% confidence intervals are indicated by the ends of the vertical bars. Each panel shows the effect sizes for either pooled data or individual biological replicates from data shown in the main figures. The numbering of figure panels corresponds to the main figures.
41586_2020_2699_MOESM3_ESM.xlsx
Supplementary Table 1 DESeq2 results comparing annotated small RNAs isolated from PA14 bacteria grown at 25 °C or 15 °C on plates. Complete DESeq2 results are reported. All differentially expressed genes (DESeq2, adjusted p-value < 0.05) are shown separately. In addition, genes upregulated at 25 °C compared to 15 °C and a summary of overlapping targets are described. N = 6 biological replicates for 25 °C, and 4 biological replicates for 15°C growth conditions.
41586_2020_2699_MOESM4_ESM.xlsx
Supplementary Table 2 DESeq2 results comparing annotated small RNAs isolated from PA14 bacteria grown at 25 °C on plated or in liquid. Complete DESeq2 results are reported. All differentially expressed genes (DESeq2, adjusted p-value < 0.05) are shown separately. In addition, genes upregulated at 25°C compared to liquid growth conditions are provided. N = 6 biological replicates for 25°C, and 4 biological replicates for liquid growth conditions.
41586_2020_2699_MOESM5_ESM.xlsx
Supplementary Table 3 P11 homology to C. elegans coding and noncoding RNAs. PA14 P11 homology with worm sequences was determined by BLAST. Results are shown for homology with worm mRNAs, piRNAs, and additional ncRNAs annotated on Wormbase.
41586_2020_2699_MOESM6_ESM.xlsx
Supplementary Table 4 Statistics reporting. Statistics were generated using Prism 8. Detailed results from all figures are provided.
Rights and permissions
About this article
Cite this article
Kaletsky, R., Moore, R.S., Vrla, G.D. et al. C. elegans interprets bacterial non-coding RNAs to learn pathogenic avoidance. Nature 586, 445–451 (2020). https://doi.org/10.1038/s41586-020-2699-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2699-5
This article is cited by
-
ADARs regulate cuticle collagen expression and promote survival to pathogen infection
BMC Biology (2024)
-
Inheritance of associative memories and acquired cellular changes in C. elegans
Nature Communications (2023)
-
The hunger strikes back: an epigenetic memory for autophagy
Cell Death & Differentiation (2023)
-
An intestinal sphingolipid confers intergenerational neuroprotection
Nature Cell Biology (2023)
-
Engram neurons: Encoding, consolidation, retrieval, and forgetting of memory
Molecular Psychiatry (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.