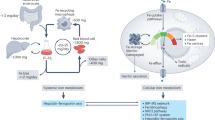
Abstract
The serum level of iron in humans is tightly controlled by the action of the hormone hepcidin on the iron efflux transporter ferroportin. Hepcidin regulates iron absorption and recycling by inducing the internalization and degradation of ferroportin1. Aberrant ferroportin activity can lead to diseases of iron overload, such as haemochromatosis, or iron limitation anaemias2. Here we determine cryogenic electron microscopy structures of ferroportin in lipid nanodiscs, both in the apo state and in complex with hepcidin and the iron mimetic cobalt. These structures and accompanying molecular dynamics simulations identify two metal-binding sites within the N and C domains of ferroportin. Hepcidin binds ferroportin in an outward-open conformation and completely occludes the iron efflux pathway to inhibit transport. The carboxy terminus of hepcidin directly contacts the divalent metal in the ferroportin C domain. Hepcidin binding to ferroportin is coupled to iron binding, with an 80-fold increase in hepcidin affinity in the presence of iron. These results suggest a model for hepcidin regulation of ferroportin, in which only ferroportin molecules loaded with iron are targeted for degradation. More broadly, our structural and functional insights may enable more targeted manipulation of the hepcidin–ferroportin axis in disorders of iron homeostasis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this Article and its Supplementary Information. Crystallographic coordinates and structure factors for the Fab45D8 complex have been deposited in the Protein Data Bank (PDB) under accession code 6W4V. Coordinates for the Fab45D8–FPN complex have been deposited in the PDB under accession code 6W4S and the maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession code 21539. Coordinates for the FPN–Co2+–hepcidin–Fab45D8 complex have been deposited in the PDB under accession code 6WBV and the maps have been deposited in the EMDB under accession code 21599.
References
Nemeth, E. et al. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science 306, 2090–2093 (2004).
Drakesmith, H., Nemeth, E. & Ganz, T. Ironing out ferroportin. Cell Metab. 22, 777–787 (2015).
Donovan, A. et al. The iron exporter ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab. 1, 191–200 (2005).
Donovan, A. et al. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature 403, 776–781 (2000).
Knutson, M. D., Oukka, M., Koss, L. M., Aydemir, F. & Wessling-Resnick, M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc. Natl Acad. Sci. USA 102, 1324–1328 (2005).
Ward, D. M. & Kaplan, J. Ferroportin-mediated iron transport: expression and regulation. Biochim. Biophys. Acta 1823, 1426–1433 (2012).
Aschemeyer, S. et al. Structure-function analysis of ferroportin defines the binding site and an alternative mechanism of action of hepcidin. Blood 131, 899–910 (2018).
De Domenico, I. et al. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol. Biol. Cell 18, 2569–2578 (2007).
Qiao, B. et al. Hepcidin-induced endocytosis of ferroportin is dependent on ferroportin ubiquitination. Cell Metab. 15, 918–924 (2012).
Ross, S. L. et al. Molecular mechanism of hepcidin-mediated ferroportin internalization requires ferroportin lysines, not tyrosines or JAK-STAT. Cell Metab. 15, 905–917 (2012).
Roetto, A. et al. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat. Genet. 33, 21–22 (2003).
De Domenico, I. et al. The molecular basis of ferroportin-linked hemochromatosis. Proc. Natl Acad. Sci. USA 102, 8955–8960 (2005).
Drakesmith, H. et al. Resistance to hepcidin is conferred by hemochromatosis-associated mutations of ferroportin. Blood 106, 1092–1097 (2005).
Roy, C. N. et al. Hepcidin antimicrobial peptide transgenic mice exhibit features of the anemia of inflammation. Blood 109, 4038–4044 (2007).
Ganz, T. & Nemeth, E. The hepcidin-ferroportin system as a therapeutic target in anemias and iron overload disorders. Hematology 2011, 538–542 (2011).
Manolova, V. et al. Oral ferroportin inhibitor ameliorates ineffective erythropoiesis in a model of β-thalassemia. J. Clin. Invest. 130, 491–506 (2019).
Witcher, D. R. et al. LY2928057, an antibody targeting ferroportin, is a potent inhibitor of hepcidin activity and increases iron mobilization in normal cynomolgus monkeys. Blood 122, 3433 (2013).
Crielaard, B. J., Lammers, T. & Rivella, S. Targeting iron metabolism in drug discovery and delivery. Nat. Rev. Drug Discov. 16, 400–423 (2017).
Vlasveld, L. T. et al. Twenty years of ferroportin disease: a review or an update of published clinical, biochemical, molecular, and functional features. Pharmaceuticals 12, 132 (2019).
Nemeth, E. et al. The N-terminus of hepcidin is essential for its interaction with ferroportin: structure-function study. Blood 107, 328–333 (2006).
Jordan, J. B. et al. Hepcidin revisited, disulfide connectivity, dynamics, and structure. J. Biol. Chem. 284, 24155–24167 (2009).
Bonaccorsi di Patti, M. C. et al. A structural model of human ferroportin and of its iron binding site. FEBS J. 281, 2851–2860 (2014).
Ross, S. L. et al. Identification of antibody and small molecule antagonists of ferroportin-hepcidin interaction. Front. Pharmacol. 8, 838 (2017).
Wu, S. et al. Fabs enable single particle cryoEM studies of small proteins. Structure 20, 582–592 (2012).
Nasr, M. L. et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat. Methods 14, 49–52 (2017).
Taniguchi, R. et al. Outward- and inward-facing structures of a putative bacterial transition-metal transporter with homology to ferroportin. Nat. Commun. 6, 8545 (2015).
Deshpande, C. N. et al. Calcium is an essential cofactor for metal efflux by the ferroportin transporter family. Nat. Commun. 9, 3075 (2018).
Guellec, J. et al. Molecular model of the ferroportin intracellular gate and implications for the human iron transport cycle and hemochromatosis type 4A. FASEB J. 33, 14625–14635 (2019).
Sham, R. L. et al. Autosomal dominant hereditary hemochromatosis associated with a novel ferroportin mutation and unique clinical features. Blood Cells Mol. Dis. 34, 157–161 (2005).
Schimanski, L. M. et al. In vitro functional analysis of human ferroportin (FPN) and hemochromatosis-associated FPN mutations. Blood 105, 4096–4102 (2005).
Viprakasit, V. et al. Molecular diagnosis of the first ferroportin mutation (C326y) in the Far East causing a dominant form of inherited iron overload. Blood 104, 3204 (2004).
Mayr, R. et al. Identification of mutations in SLC40A1 that affect ferroportin function and phenotype of human ferroportin iron overload. Gastroenterology 140, 2056–2063 (2011).
Tortosa, V., di Patti, M. C. B., Brandi, V., Musci, G. & Polticelli, F. An improved structural model of the human iron exporter ferroportin. Insight into the role of pathogenic mutations in hereditary hemochromatosis type 4. Bio-Algorithms Med-Systems 13, 215–222 (2017).
Wallace, D. F., Clark, R. M., Harley, H. A. J. & Subramaniam, V. N. Autosomal dominant iron overload due to a novel mutation of ferroportin1 associated with parenchymal iron loading and cirrhosis. J. Hepatol. 40, 710–713 (2004).
Njajou, O. T. et al. A mutation in SLC11A3 is associated with autosomal dominant hemochromatosis. Nat. Genet. 28, 213–214 (2001).
Arden, K. E. et al. A novel mutation in ferroportin1 is associated with haemochromatosis in a Solomon Islands patient. Gut 52, 1215–1217 (2003).
Zhang, W. et al. A novel SLC40A1 p.Y333H mutation with gain of function of ferroportin: a recurrent cause of haemochromatosis in China. Liver Int. 39, 1120–1127 (2019).
Létocart, E. et al. A novel missense mutation in SLC40A1 results in resistance to hepcidin and confirms the existence of two ferroportin-associated iron overload diseases. Br. J. Haematol. 147, 379–385 (2009).
Callebaut, I. et al. Comprehensive functional annotation of 18 missense mutations found in suspected hemochromatosis type 4 patients. Hum. Mol. Genet. 23, 4479–4490 (2014).
Preza, G. C. et al. Minihepcidins are rationally designed small peptides that mimic hepcidin activity in mice and may be useful for the treatment of iron overload. J. Clin. Invest. 121, 4880–4888 (2011).
Rivard, S. R. et al. Autosomal dominant reticuloendothelial iron overload (HFE type 4) due to a new missense mutation in the FERROPORTIN 1 gene (SLC11A3) in a large French-Canadian family. Haematologica 88, 824–826 (2003).
Raszeja-Wyszomirska, J., Caleffi, A., Milkiewicz, P. & Pietrangelo, A. Ferroportin-related haemochromatosis associated with novel Y64H mutation of the SCL40A1 gene. Prz. Gastroenterol. 9, 307–309 (2014).
Galesloot, T. E. et al. Serum hepcidin: reference ranges and biochemical correlates in the general population. Blood 117, e218–e225 (2011).
Winter, G., Lobley, C. M. C. & Prince, S. M. Decision making in xia2. Acta Crystallogr. D 69, 1260–1273 (2013).
Kabsch, W. XDS. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Aleman, F. et al. Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors. Proc. Natl Acad. Sci. USA 115, 7569–7574 (2018).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Cote, L. J., Kim, F. & Huang, J. Langmuir-Blodgett assembly of graphite oxide single layers. J. Am. Chem. Soc. 131, 1043–1049 (2009).
Palovcak, E. et al. A simple and robust procedure for preparing graphene-oxide cryo-EM grids. J. Struct. Biol. 204, 80–84 (2018).
Wang, F. et al. Amino and PEG-amino graphene oxide grids enrich and protect samples for high-resolution single particle cryo-electron microscopy. J. Struct. Biol. 209, 107437 (2020).
Mastronarde, D. N. & Serial, E. M. A Program for automated tilt series acquisition on tecnai microscopes using prediction of specimen position. Microsc. Microanal. 9, 1182–1183 (2003).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
de la Rosa-Trevín, J. M. et al. Scipion: A software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Asarnow, D., Palovcak, E. & Cheng, Y. asarnow/pyem: UCSF pyem v0.5. https://doi.org/10.5281/zenodo.3576630 (2019).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Dang, S. et al. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552, 426–429 (2017).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol. 184, 226–236 (2013).
Eswar, N. et al. Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. Ch. 2, Unit 2.9 (2007).
Goddard, T. D., Huang, C. C. & Ferrin, T. E. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 157, 281–287 (2007).
Barad, B. A. et al. EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946 (2015).
Betz, R. Dabble. https://doi.org/10.5281/zenodo.836914 (2017).
Olsson, M. H. M., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Søndergaard, C. R., Olsson, M. H. M., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput. 7, 2284–2295 (2011).
Morozenko, A. & Stuchebrukhov, A. A. Dowser++, a new method of hydrating protein structures. Proteins 84, 1347–1357 (2016).
Lomize, M. A., Lomize, A. L., Pogozheva, I. D. & Mosberg, H. I. OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 (2006).
Klauda, J. B. et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010).
Huang, J. et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods 14, 71–73 (2017).
Beglov, D. & Roux, B. Finite representation of an infinite bulk system: Solvent boundary potential for computer simulations. J. Chem. Phys. 100, 9050–9063 (1994).
Li, P., Roberts, B. P., Chakravorty, D. K. & Merz, K. M. Jr. Rational design of particle mesh Ewald compatible Lennard-Jones parameters for+ 2 metal cations in explicit solvent. J. Chem. Theory Comput. 9, 2733–2748 (2013).
Case, D. A. et al. AMBER University of California, San Francisco. https://ambermd.org/index.php (2018).
Salomon-Ferrer, R., Götz, A. W., Poole, D., Le Grand, S. & Walker, R. C. Routine microsecond molecular dynamics simulations with AMBER on gpus. 2. explicit solvent particle mesh ewald. J. Chem. Theory Comput. 9, 3878–3888 (2013).
Hopkins, C. W., Le Grand, S., Walker, R. C. & Roitberg, A. E. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 11, 1864–1874 (2015).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Roe, D. R. & Cheatham, T. E. III. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 9, 3084–3095 (2013).
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–88 (1996).
Mitchell, C. J., Shawki, A., Ganz, T., Nemeth, E. & Mackenzie, B. Functional properties of human ferroportin, a cellular iron exporter reactive also with cobalt and zinc. Am. J. Physiol. Cell Physiol. 306, C450–C459 (2014).
Acknowledgements
We thank Zanlin Yu for assistance with cryo-EM grid preparation and data collection. This work was supported by National Institutes of Health (NIH) grants DP5OD023048 (A.M.), R01GM098672 (Y.C.), and P50AI150476 (Y.C.). A.S.P. was supported by a National Science Foundation Graduate Research Fellowship and NIH training grant T32GM120007. R.C.K. was supported by NIH training grant T32GM008293. Cryo-EM equipment at UCSF is partially supported by NIH grants S10OD020054 and S10OD021741. C.B.B. was supported by the Alfred Benzon Foundation. R.O.D. acknowledges support from a Stanford University Bio-X seed grant. Y.C. is an Investigator of Howard Hughes Medical Institute. A.M. acknowledges support from the Pew Charitable Trusts, the Esther and A. & Joseph Klingenstein Fund and the Searle Scholars Program.
Author information
Authors and Affiliations
Contributions
C.B.B. purified FPN and Fab fragments, established biochemical approaches to reconstitute a FPN–hepcidin complex, established and performed hepcidin binding and calcein transport assays, prepared samples for cryo-EM, screened samples by negative stain and cryo-EM, and determined the X-ray crystal structure of Fab45D8. C.M.A. identified optimal freezing conditions for FPN samples, collected cryo-EM data, and determined high resolution cryo-EM maps by extensive image processing. R.C.K. and A.S.P. performed and analysed molecular dynamics simulations under the guidance of R.O.D. S.G. collected negative-stain and cryo-EM data for FPN samples bound to different Fabs, and identified Fab45D8 as a fiducial marker for structure determination. S.S. aided in establishing and performing the calcein transport assay. T.A. provided fluorescently labelled hepcidin and anti-FPN antibodies for structure determination. A.M. built and refined models of FPN and Fab45D8. The manuscript was written by C.B.B., C.M.A., Y.C. and A.M. with revisions provided by all authors. The overall project was supervised by Y.C. and A.M.
Corresponding authors
Ethics declarations
Competing interests
T.A. is employed by Amgen and reports Amgen stock. The other authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Extended data figures and tables
Extended Data Fig. 1 Biochemistry of purified human ferroportin and Fab45D8 complex.
a, Size exclusion chromatography of purified apo- and hepcidin–Co2+-bound FPN–Fab45D8 complex. SDS–PAGE gel of purified complex under non-reducing (NR) and reducing (R) conditions. SEC and SDS–PAGE analyses are representative and were performed four and two times for apo- and hepcidin–Co2+-bound FPN–Fab45D8 samples respectively. b, Fab45D8 binds nanodisc-reconstituted FPN (nanodisc–FPN), as assessed by size-exclusion chromatography. c, Fluorescence size exclusion chromatography (FSEC) of RhoG–hepcidin. RhoG–hepcidin co-elutes with nanodisc–FPN and with a nanodisc–FPN–Fab45D8 complex in the presence of CoCl2. d, FSEC shows RhoG–hepcidin binding to nanodisc–FPN is competed by excess unlabelled hepcidin used for structural studies. e, Fluorescence polarization of RhoG–hepcidin increases further with Fab45D8, consistent with formation of a larger complex. Importantly, Fab45D8 does not decrease RhoG–hepcidin binding. Data points are means from n = 3 technical replicates. f, RhoG–hepcidin binding to nanodisc-FPN in the presence of 10 μM CoCl2 measured by fluorescence polarization in the absence (Kd = 31 nM) and presence of 3 μM Fab45D8 (Kd = 39 nM). Data points are means from n = 3 technical replicates. g, RhoG–hepcidin binds to FPN (FPN + hepcidin) but not to Fab45D8 alone (Fab45D8 + hepcidin). The order of hepcidin and Fab45D8 addition does not influence the increase in fluorescence polarization for a Fab45D8–FPN–hepcidin complex. Data points are means from n = 4 technical replicates.
Extended Data Fig. 2 Calcein-quenching assay for FPN metal transport.
a, Schematic of calcein-based assay to measure transport of divalent cations. Purified FPN is reconstituted into proteoliposomes containing calcein. Transport of divalent cations leads to quenching of calcein fluorescence, which can also occur through the divalent cation-selective ionophore calcimycin. b, c, Specificity of the calcein transport assay. FPN-reconstituted liposomes facilitate uptake of 1,000 μM Co2+ added (arrows, red trace) to the external bath solution, producing a calcein-quenching response that follows a single exponential. Further additions of Co2+ demonstrates the transport process has reached saturation. Calcein fluorescence from empty liposomes (black trace) returns to baseline after multiple additions of 1,000 μM Co2+. c, Additions of Co2+ followed by calcimycin results in rapid and almost complete quenching. Fluorescence traces are representative experiments. d, Liposome reconstitution samples analysed by reducing SDS–PAGE, the analysis is representative and was performed three times for FPN samples and twice for FPN samples containing FPN–hepcidin or FPN–Fab45D8. e, Calcein transport assay queried against a panel of putative transition metal ion substrates at 100 μM. Notably, the larger ions Ni2+ and Zn2+ are also reported FPN substrates83. The smaller ion Mn2+ was previously found not to be effluxed by FPN in oocytes83, but appears to be transported in liposomes. Cu2+ interferes with empty and FPN-containing liposomes, leading to non-specific quenching of calcein. Fluorescence traces are representative experiments. f, Fab45D8 decreases cobalt transport Vmax (−2.40 × 10−3 ± 0.075 × 10−3 ΔF s−1) and Km (4.68 ± 0.54 μM) values. Data are mean ± s.e.m. from n = 3 independent experiments.
Extended Data Fig. 3 Cryo-EM data processing for FPN–Fab45D8.
a, Representative motion-corrected micrograph collected on the Titan Krios showing monodisperse FPN–Fab45D8 nanodisc particles. Four different FPN–Fab45D8 samples were imaged on GO-coated Quantifoil grids times with similar results. b, Examples of ‘good’ 2D class averages that were used in 3D classification. Similar quality class averages were produced when processing six unique subsets of the final particles used. Scale bar, 50 Å. c, Flowchart showing image processing pipeline for FPN–Fab45D8. Initial processing, through 2D classification, was performed in cryoSPARC. Particles were then transferred, using csparc2star.py, to RELION for 3D classification, then to cryoSPARC for a nonuniform refinement, and finally to cisTEM for refinement. The number of particles moving into each step is noted. d, Final refinements from cryosparc and cisTEM beside their directional FSC curves calculated using dfsc.0.0.1.py. Angular distribution plot from cisTEM is shown.
Extended Data Fig. 4 Cryo-EM data processing for Co2+–hepcidin–FPN–Fab45D8.
a, Representative motion-corrected micrograph collected on the Titan Krios showing monodisperse Co2+–hepcidin–FPN–Fab45D8 nanodisc particles. Two independent Co2+–hepcidin–FPN–Fab45D8 samples were frozen on UltrAufoil grids with similar micrograph quality. b, Examples of ‘good’ 2D class averages that were used in 3D classification. c, Flowchart showing image processing pipeline for Co2+–hepcidin–FPN–Fab45D8. Initial processing was performed in cryoSPARC. Local CTF refinement, with a conservative high-resolution limit of 4 Å, was performed before nonuniform refinement using default parameters. A final subset of particles was transferred to cisTEM for refinement. The number of particles moving into each step is noted. d, Final refinements from cryosparc and cisTEM beside their directional FSC curves calculated using dfsc.0.0.1.py. Angular distribution plot from cisTEM is also shown.
Extended Data Fig. 5 Cryo-EM map density for FPN.
a, c, Local resolution estimation in cryoSPARC for the apo-FPN–Fab45D8 complex (a) and for the Co2+–hepcidin–FPN–Fab45D8 complex (c). Two views of each complex are shown. b, d, Shown in grey mesh is cryo-EM map density for individual FPN transmembrane helices for the apo-FPN–Fab45D8 complex (b) and for the Co2+–hepcidin–FPN–Fab45D8 complex (d). Mesh depicts density within a 2.5 Å radius of any modelled atom.
Extended Data Fig. 6 Structure of Fab45D8 and comparison of human FPN with BbFPN.
a, Cryo-EM density of FPN and Fab45D8. Although density is observed for the constant regions of Fab45D8, it is not of sufficient quality to unambiguously model. b, Fab45D8 makes extensive contacts with FPN ECL2 with both the heavy (VH) and light (VL) chains. c, Crystal structure of Fab45D8 at 2.1 Å. d, Comparison of Fab45D8 alone (transparent cartoon and sticks) and bound to FPN. The binding site residues of Fab45D8 change minimally upon binding FPN. e, Human FPN aligned to the outward-open (PDB code 5AYN) and inward-open (PDB code 5AYO) conformations of BbFPN. f, Unique architecture of TM7 shared between human FPN and BbFPN.
Extended Data Fig. 7 Effect of pH and ions on metal transport and binding.
a, b, Co2+ transport by FPN is potentiated by an opposing H+-gradient. a, Time-course analysis of calcein fluorescence-quenching response after addition of 300 μM CoCl2 (arrow) for FPN–liposomes containing 100 mM KCl and HEPES, pH 7.5 that were diluted into buffer consisting of 100 mM NaCl and (i) HEPES pH 6.8, (ii) HEPES pH 7.5, (iii) HEPES pH 8.2, (iv) Tris/MES pH 6.0, or (v) Tris/MES pH 9.0. b, Initial rates of transport obtained from the linear phase of time-course fluorescence-quenching experiments at the indicated pH values. Data are mean ± s.e.m. from n = 3 experiments. c, Calcein fluorescence responses for FPN–liposomes containing 100 mM KCl and HEPES, pH 7.5, diluted into 100 mM NaCl and HEPES, pH 7.5. A membrane potential (ΔΨ) was generated by addition of the K+ selective ionophore valinomycin before the experiment, which creates an outward K+ gradient. A non-significant (P = 0.5986, Student’s unpaired t-test, two-tailed) difference was observed for the rate of transport by valinomycin addition. Data are mean ± s.e.m. with corresponding data points from n = 3 independent experiments. d–g, Calcium modulates the FPN transport mechanism. d, Time-course analysis of calcein fluorescence-quenching response after addition of 1,000 μM CoCl2 (arrow) for FPN–liposomes preloaded with 1.25 mM CaCl2 internally (red) or 1.25 mM CaCl2 added to the external solution (grey). e, Initial rates of Co2+ transport in the presence of internal 1.25 mM CaCl2 (red circles). Data are mean ± s.e.m. from n = 3 independent experiments. Quantified transport rates are in Supplementary Table 2. f, Calcein fluorescence response for FeCl2. g, Initial rates of Fe2+ transport. Data are mean ± s.e.m. from n = 3 independent experiments. h, i, Calcium does not support high-affinity hepcidin binding to FPN. h, Binding of 5 nM RhoG–hepcidin to nanodisc–FPN in the presence of 3 mM CaCl2 was determined by fluorescence polarization (EC50 > 100 nM). Data are mean ± s.e.m. from n = 3 independent experiments. i, Ca2+ stimulates the partial binding of 5 nM RhoG–hepcidin to 100 nM nanodisc–FPN with an EC50 of 47.1 ± 4.1 μM, confirming that the Ca2+-binding site in FPN titrates in the physiological range (approximately 1.25 mM) of free ionized Ca2+. By contrast, Co2+ stimulates binding of 5 nM RhoG–hepcidin to 100 nM nanodisc–FPN with an EC50 of 35.8 ± 3.6 nM. However, depletion of RhoG–hepcidin leads to a nHill value of 0.52, and the true affinity of cobalt may be higher than this value. Notably, these results indicate that binding of physiologically relevant concentrations of hepcidin would be exceedingly ineffective to apo- and Ca2+-bound FPN. Data are mean ± s.e.m. from n = 3 independent experiments.
Extended Data Fig. 8 Molecular dynamics simulations of iron binding to apo-FPN.
a, Local resolution of FPN cryo-EM density. Insets show local resolution around TM7b. b, Each graph corresponds to an independent simulation in which Fe2+ ions start in solution and bind spontaneously to the proposed iron binding site. Distance shown is from the ion to the nearest oxygen atom of the D325 side chain. Thick traces represent a 15-ns sliding mean and thin traces represent unsmoothed values. Time traces include 30 ns of equilibration. The average time to bind (the time from start of production simulation to when the measured distance is less) was 71 ns. c, Representative conformations of TM7 from simulations with iron bound (blue) or absent (orange). d, In the absence of bound iron, D325 is mobile and can move into the cavity between TM7b and TM1. The top of TM7b can also tilt away from TM1. Four representative frames from simulation are overlaid. e, In simulations with Fe2+ bound, interaction with the ion restricts the mobility of D325 and, in turn, TM7b. f, Iron-bound and iron-absent simulations show differences in the dynamics and position of D325, as measured by the distance between D325 Cγ and S47 Cβ. g, Comparison of conformation and dynamics with and without iron bound. With iron bound, D325 moves away from TM1 into the iron binding site (left), the root-mean-square fluctuation (RMSF) of D325 decreases (middle), and the extracellular end of TM7b moves closer to TM1 (right). For these comparisons, six simulations for each condition were used, each 2.0 μs in length. Error bars are s.e.m. P = 0.001 (left), P = 0.046 (middle) and P = 0.046 (right), two-tailed Mann–Whitney U test.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1 and 2. These include a table summarizing molecular dynamics simulations and a table summarizing kinetics data from the calcein assay.
Rights and permissions
About this article
Cite this article
Billesbølle, C.B., Azumaya, C.M., Kretsch, R.C. et al. Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms. Nature 586, 807–811 (2020). https://doi.org/10.1038/s41586-020-2668-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2668-z
This article is cited by
-
Iron imbalance in neurodegeneration
Molecular Psychiatry (2024)
-
Mechanisms controlling cellular and systemic iron homeostasis
Nature Reviews Molecular Cell Biology (2024)
-
Ferritinophagy induced ferroptosis in the management of cancer
Cellular Oncology (2024)
-
Ferroptosis regulation through Nrf2 and implications for neurodegenerative diseases
Archives of Toxicology (2024)
-
Apo- and holo-transferrin differentially interact with hephaestin and ferroportin in a novel mechanism of cellular iron release regulation
Journal of Biomedical Science (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.