Abstract
The G-protein-coupled bile acid receptor (GPBAR) conveys the cross-membrane signalling of a vast variety of bile acids and is a signalling hub in the liver–bile acid–microbiota–metabolism axis1,2,3. Here we report the cryo-electron microscopy structures of GPBAR–Gs complexes stabilized by either the high-affinity P3954 or the semisynthesized bile acid derivative INT-7771,3 at 3 Å resolution. These structures revealed a large oval pocket that contains several polar groups positioned to accommodate the amphipathic cholic core of bile acids, a fingerprint of key residues to recognize diverse bile acids in the orthosteric site, a putative second bile acid-binding site with allosteric properties and structural features that contribute to bias properties. Moreover, GPBAR undertakes an atypical mode of activation and G protein coupling that features a different set of key residues connecting the ligand-binding pocket to the Gs-coupling site, and a specific interaction motif that is localized in intracellular loop 3. Overall, our study not only reveals unique structural features of GPBAR that are involved in bile acid recognition and allosteric effects, but also suggests the presence of distinct connecting mechanisms between the ligand-binding pocket and the G-protein-binding site in the G-protein-coupled receptor superfamily.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The cryo-EM density map for the P395–GPBAR–Gs complex and the INT-777–GPBAR–Gs complex have been deposited in the EMDB under the accession codes EMD-30344 and EMD-30345, respectively. The coordinates for the model of P395–GPBAR–Gs and INT-777–GPBAR–Gs have been deposited in the PDB under the accession numbers 7CFM and 7CFN, respectively. All other data are available on request from the corresponding authors.
References
Jia, W., Xie, G. & Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018).
McGavigan, A. K. et al. TGR5 contributes to glucoregulatory improvements after vertical sleeve gastrectomy in mice. Gut 66, 226–234 (2017).
Perino, A. & Schoonjans, K. TGR5 and immunometabolism: insights from physiology and pharmacology. Trends Pharmacol. Sci. 36, 847–857 (2015).
Piotrowski, D. W. et al. Identification of tetrahydropyrido[4,3-d]pyrimidine amides as a new class of orally bioavailable TGR5 agonists. ACS Med. Chem. Lett. 4, 63–68 (2012).
Qi, X. et al. Gut microbiota–bile acid–interleukin-22 axis orchestrates polycystic ovary syndrome. Nat. Med. 25, 1225–1233 (2019).
Watanabe, M. et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 439, 484–489 (2006).
Schaap, F. G., Trauner, M. & Jansen, P. L. Bile acid receptors as targets for drug development. Nat. Rev. Gastroenterol. Hepatol. 11, 55–67 (2014).
Pols, T. W., Noriega, L. G., Nomura, M., Auwerx, J. & Schoonjans, K. The bile acid membrane receptor TGR5 as an emerging target in metabolism and inflammation. J. Hepatol. 54, 1263–1272 (2011).
Guo, C. et al. Cholesterol homeostatic regulator SCAP–SREBP2 integrates NLRP3 inflammasome activation and cholesterol biosynthetic signaling in macrophages. Immunity 49, 842–856.e7 (2018).
Perino, A. et al. TGR5 reduces macrophage migration through mTOR-induced C/EBPβ differential translation. J. Clin. Invest. 124, 5424–5436 (2014).
Velazquez-Villegas, L. A. et al. TGR5 signalling promotes mitochondrial fission and beige remodelling of white adipose tissue. Nat. Commun. 9, 245 (2018).
Thomas, C. et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 10, 167–177 (2009).
Hu, M. M. et al. Virus-induced accumulation of intracellular bile acids activates the TGR5–β-arrestin–SRC axis to enable innate antiviral immunity. Cell Res. 29, 193–205 (2019).
Latorraca, N. R., Venkatakrishnan, A. J. & Dror, R. O. GPCR dynamics: structures in motion. Chem. Rev. 117, 139–155 (2017).
Isberg, V. et al. Generic GPCR residue numbers—aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 36, 22–31 (2015).
García-Nafría, J., Lee, Y., Bai, X., Carpenter, B. & Tate, C. G. Cryo-EM structure of the adenosine A2A receptor coupled to an engineered heterotrimeric G protein. eLife 7, e35946 (2018).
Rosenbaum, D. M. et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science 318, 1266–1273 (2007).
Martin, R. E. et al. 2-Phenoxy-nicotinamides are potent agonists at the bile acid receptor GPBAR1 (TGR5). ChemMedChem 8, 569–576 (2013).
Rajagopal, S. et al. Quantifying ligand bias at seven-transmembrane receptors. Mol. Pharmacol. 80, 367–377 (2011).
Liang, Y. L. et al. Phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor–Gs complex. Nature 555, 121–125 (2018).
Cleator, J. H., Mehta, N. D., Kurtz, D. T. & Hildebrandt, J. D. The N54 mutant of Gαs has a conditional dominant negative phenotype which suppresses hormone-stimulated but not basal cAMP levels. FEBS Lett. 443, 205–208 (1999).
Iiri, T., Bell, S. M., Baranski, T. J., Fujita, T. & Bourne, H. R. A Gsα mutant designed to inhibit receptor signaling through Gs. Proc. Natl Acad. Sci. USA 96, 499–504 (1999).
Lee, E., Taussig, R. & Gilman, A. G. The G226A mutant of Gs alpha highlights the requirement for dissociation of G protein subunits. J. Biol. Chem. 267, 1212–1218 (1992).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Heymann, J. B. Guidelines for using Bsoft for high resolution reconstruction and validation of biomolecular structures from electron micrographs. Protein Sci. 27, 159–171 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D Struct. Biol. 74, 814–840 (2018).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
The PyMOL Molecular Graphics System, Version 2.0 (Schrödinger, 2017).
Stoddart, L. A. et al. Application of BRET to monitor ligand binding to GPCRs. Nat. Methods 12, 661–663 (2015).
Fan, F. et al. Novel genetically encoded biosensors using firefly luciferase. ACS Chem. Biol. 3, 346–351 (2008).
Hu, Q. X. et al. Constitutive Gαi coupling activity of very large G protein-coupled receptor 1 (VLGR1) and its regulation by PDZD7 protein. J. Biol. Chem. 289, 24215–24225 (2014).
Yang, F. et al. Allosteric mechanisms underlie GPCR signaling to SH3-domain proteins through arrestin. Nat. Chem. Biol. 14, 876–886 (2018).
Lu, J. et al. Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40. Nat. Struct. Mol. Biol. 24, 570–577 (2017).
Acknowledgements
We acknowledge support from the National Key R&D Program of China (grants 2018YFC1003600 to X.Y. and J.-P.S., and 2019YFA050880 to Y.Z. and P.X.), the National Science Fund for Excellent Young Scholars (grants 81822008 to X.Y. and 81922071 to Y.Z.), the National Science Fund for Distinguished Young Scholars (grants 81773704 to J.-P.S., 81425024 to X.X., and 81525005 to F.Yi.), Zhejiang Province National Science Fund for Excellent Young Scholars (grant LR19H310001 to Y.Z.), the National Natural Science Foundation of China (grants 31900936 to F.Yang., 31700692 to P.X. and 81730099 to X.X.), the China Postdoctoral Science Foundation (grant 2019T120587 to F.Yang.), and the Innovative Research Team in University (grant IRT_17R68 to X.Y. and Y.S.). Sample preparation for cryo-EM studies was supported by the Protein Facility, Zhejiang University School of Medicine. The cryo-EM data were collected at the Center of Cryo-Electron Microscopy, Zhejiang University, with assistance from S. Chang.
Author information
Authors and Affiliations
Contributions
Y.Z., X.X., X.Y. and J.-P.S. organized the whole project. J.-P.S., Y.Z., X.Y. and X.X. supervised the overall project design and execution. X.X. provided all of the ligand study and chemical guidance. Y.Z. guided all of the cryo-EM study. Y.Z. and J.-P.S. guided all of the structural analysis. X.Y., Y.Z., J.-P.S. and X.X. designed all of the mutants for the ligand-binding pocket and the second bile acid-binding site. X.Y. initiated the study of recognition mechanism of bile acid, allosteric assays and bile acid derivatives by GPBAR and designed the screening assay for complex formation. X.Y. designed the biased signalling assay. X.Y. and J.-P.S. designed the cooperative assay for the allosteric mechanism. X.X., C.Z. and F.N. designed and synthesized the fluorescent-tagged ligand for the binding assay. J.-P.S., X.Y. and F. Yang designed the ligand-binding assay. J.-P.S. and X.Y. designed all of the cellular experimental details. F. Yang, L.G. and P.X. developed the GPBAR constructs and optimized protein expression. F. Yang, L.G. and P.X. established the P395–GPBAR–Gs and INT-777–GPBAR–Gs complex formation strategy. F. Yang, L.G., P.X. and X.W. screened the bile acids or its derivatives for complex formation. F. Yang, L.G., P.X. and X.W. performed virus production, insect cell expression and prepared samples for cryo-EM. F. Yang and L.G. developed the method for solubilization of the bile acids advised by Y.Z., J.-P.S. and X.Y. F. Yang and L.G. screened conditions for gel filtration. D.-D.S. evaluated the sample by negative-stain EM. C. Mao prepared the cryo-EM grids. C. Mao and D.-D.S. collected the cryo-EM data with assistance from Q.S. C.Mao and Q.M. performed cryo-EM map calculation, model building and structure refinement. Q.S. performed the MD simulations. C.Ma. assisted in protein purification in the Protein Facility. L.G., P.X. and K.Z. performed the pull-down assay. J.L. performed the cooperative assay and data analysis. X.W., L.G., J.L., S.G., L.Z., F.Yi., Y.P., X.L. and K.Z. performed the cAMP accumulation assay and binding assay. X.W., Y.P., R.L. and S.G. performed the BRET assay. Y.S., F.Yi., J.Z. and C.J. participated in the design and explanation of the cAMP and BRET results and provided insightful ideas and experimental designs. V.C.L. oversaw the structural analysis. J.-P.S. wrote the manuscript. All the authors have seen and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks David D. Moore and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Cryo-EM images and single-particle reconstruction of the P395–GPBAR–Gs and INT-777–GPBAR–Gs complex.
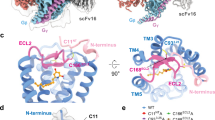
a, Left panel: representative elution profile of Flag-purified P395–GPBAR–Gs complex on Superose 6 Increase 10/30 column and SDS–PAGE of the size-exclusion chromatography peak. Right panel, coomassie-stained PAGE of the isolated perk fraction from the Superose 6. Representative Figures from three independent experiments was shown. Full-length human GPBAR with thermostabilized cytochrome b562RIL (BRIL) introduced into the N terminus was coexpressed with Gs protein in Spodoptera frugiperda (Sf9) insect cells. Active complexes were readily formed by the addition of excess high-affinity agonist P395 and the nanobody Nb35. b, Effects of different bile acids and bile acid derivatives on GPBAR–Gs complex formation, using P395 as positive control. The low solubility and affinity of endogenous bile acids complicate the formation of bile acid–GPBAR–Gs complexes in vitro. We screened a panel of bile acids and identified that only INT-777 robustly promoted a high fraction of GPBAR–Gs complex formation. Western blot of Flag-GPBAR, Gs were detected by specific antibody and quantified using densitometry. Representative western blots from at least three independent experiments were shown. (For raw data, see Supplementary Fig. 1). c, d, Cryo-EM micrograph (Scale bar: 30 nm) and reference-free two-dimensional class averages (Scale bar: 5 nm) of the P395–GPBAR–Gs complex (c) and INT-777–GPBAR–Gs complex (d). Representative Cryo-EM micrograph from 4826 movies or 6229 movies (shown in e or f respectively) and representative two-dimensional class averages determined using approximately 0.3 million (c) or 1 million (d) particles after 3D classification were shown. e, f, Flow chart of cryo-EM data processing of P395–GPBAR–Gs complex (e) and INT-777–GPBAR–Gs complex (f). The GPBAR–Gs complexes stabilized by P395 or INT-777 were analysed by single-particle Cryo-EM, which enabled us to construct electron density maps with an overall resolution of 3.0-Å.
Extended Data Fig. 2 Selected electron microscopy density map of the P395–GPBAR–Gs complex and the INT-777–GPBAR–Gs complex.
a, 3D density map colored according to local resolution (Å) of the P395 and INT-777–bound complexes. b, Gold-standard FSC (blue) curves and the model-vs-map (green), FSCwork (yellow) and FSCfree (pink) validation curves of P395 and INT-777 bound GPBAR-Gs complexes. The marginal gap between the FSCwork and FSCfree curves indicate no over-fitting of the models. c, d, EM density of the transmembrane helices, ligands and ICL3 of GPBAR and the α5 helix of Gαs of P395-GPBAR-Gs complex (c) and INT-777–GPBAR–Gs (d) complex respectively. GPBAR including all seven transmembrane (TM) helices with both intracellular and extracellular loops (ICLs and ECLs, respectively) was confidently modelled using the high-resolution electron density, and the majority of the side chains from the receptor and the G proteins were clearly identified. In particular, a well-defined density was observed for ICL3, which is not well resolved in any of the available GPCR–Gs structures. In the receptor orthosteric binding pocket, well-defined electron densities were unambiguously assigned to the compound P395 or the bile acid derivative INT-777.
Extended Data Fig. 3 Activation of GPBAR by P395 and INT-777.
a, Diagram of P395 interaction in the ligand binding pocket of GPBAR. The hydrogen bond is depicted as a dashed line. b, The restraint of the hydrophobic pathes forces the folding of P395 into a U-shaped configuration rather than an extended topology, as predicted by previous studies. c, Schematic representation of the chemical structure of INT-777 (4 rings, ring A, B, C and D were designated). d, The INT-777 assumes a flat structure, folding between the ring A and the ring B of its steroid core with an approximately 100 degree. The ring B-D is parallel to TM2. e, The orientation of the INT-777 aligned to the TM2, TM3, TM5, TM6 or TM7 of GPBAR. f, Structural comparison of the INT-777 and the P395 at the GPBAR ligand binding pocket. Colour usage: INT-777, slate; P395, magenta; GPBAR in the INT-777–GPBAR–Gs complex, green; GPBAR in the P395–GPBAR–Gs complex, slate.
Extended Data Fig. 4 Binding, activation and mutation effects of GPBAR in response to different agonists.
a, Interaction differences in the orthosteric pocket of the GPBAR bound by INT-777 and P395. Residues participating in hydrogen bonding between the ligand and receptor are highlighted in red. Residues that interact with the ligand are indicated by green circle, or that show no interaction with ligand are indicated by blank circle. Ballesteros–Weinstein residue numbers are provided for reference. b, Effects of mutations of GPBAR ligand binding pocket on the ligand binding of P395, INT-777, CA and DCA. Mutations of L166 and E169 only affected INT-777, likely due to the ethyl group at the 6 position of INT-777. Moreover, mutations of S247A and L244A show different effects on the ligand binding between CA and DCA, which differs at (R)-OH of R1 position. Original method and data are referred to Supplementary Figs. 3, 4. Values are the mean ± SEM of three independent experiments for the wild type (WT) and mutants. Statistical differences between WT and mutations were determined by two-sided one-way ANOVA with Tukey test (*, P < 0.05; **, P < 0.01; ***, P < 0.001, n.s., no significant difference). P = 0.0028, 0.0026, 0.0005, 0.0008, 0.0054, 0.0056, 0.0511, 0.0003, 0.0075, 0.0184, 0.1799, 0.0069, 0.0003, <0.0001, 0.0133 from top to bottom for P395 group; P = <0.0001, 0.0002, 0.0371, 0.0143, <0.0001, 0.0013, 0.0005, 0.001, <0.0001, <0.0001, <0.0001, 0.0001, 0.003, 0.0082, 0.0002, <0.0001 from top to bottom for INT-777 group; P = <0.0001, <0.0001, <0.0001, 0.0171, 0.0021, 0.0007, 0.0012, 0.0005, 0.0563, 0.1398, 0.0002, 0.0083, 0.0007, 0.0009, <0.0001, <0.0001 from top to bottom for CA group; P = 0.0022, 0.0023, 0.0017, 0.0005, 0.0007, 0.0045, 0.0003, 0.0009, 0.866, 0.2, <0.0001, 0.2115, 0.1958, 0.0232, <0.0001, 0.0001 from up to bottom for DCA group. c, Effects different mutations within the ligand binding pocket of GPBAR on INT-777 and CA binding. Data from at least three independent measurements are measured as mean ± SEM. d, Key interactions between the acyl linker of the P395 and the hydrophobic cleft of the bottom of the ligand binding pocket. e, Several key residues that mediate the INT-777 induced GPBAR activation were shown. The INT-777 forms hydrogen bonds with the S247 and Y240 respectively. The ring A of INT-777 also forms hydrophobic packing with the F96 at the bottom of the ligand binding pocket. Hydrogen bonds were showed in red dash. f, g, Effects of Y240F and S247A mutations on the P395 (f) or INT-777 (g) induced cAMP accumulation. Dose responses curves were shown. Data from three independent measurements are measured as mean ± SEM.
Extended Data Fig. 5 Structural fingerprints of GPBAR recognizing different bile acids.
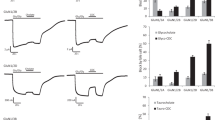
a, Diagram of the fingerprint that differentiates diverse bile acids (left panel). The 7 (R1), 12 (R2) and C-terminal (R3) positions are the most common substitution or conjugating sites in the primary bile acid CA to generate diverse bile acids, which are shown in red. Residues shown for interaction with the R1, R2 and R3 positions in GPBAR are shaded in red, green, and yellow, respectively. Substitution and conjugation status of INT-777, CA and several different bile acids at the R1, R2 and R3 positions are summarized in a table shown on the right panel. b, Diagram of the potential primary bile acid Cholic Acid (CA) interaction in the ligand binding pocket of GPBAR. Blue, residues located in the INT-777 binding pocket and shown mutating effects on both CA and INT-777; Green, residues with mutating effects only on INT-777, but not CA. c–e, Effects of bile acid recognition fingerprint mutants on cAMP accumulation induced by different bile acids. (c), mutation of L244 to A; (d), mutation of L266 to A; (e), mutation of L263 to A. The fold of EC50 change of mutant. vs. wild type for each individual bile acid were used for straightforward view. The original data were referred to Extended Data Fig. 6. Values are the mean ± SEM of three independent experiments for the wild type (WT) and mutants. Statistical differences between WT and mutations were determined by two-sided one-way ANOVA with Tukey test. *, P < 0.05, **, P < 0.01, ***, P < 0.001, n.s., no significant difference. P = <0.0001, 0.0001, 0.1796, 0.3821, 0.0002, 0.0908, 0.0127, 0.0004, 0.4761, 0.1811 from left to right for R1 group; P = <0.0001, 0.0001, 0.2056, <0.0001, 0.0694, 0.0519, 0.0089, 0.0053, <0.0001, 0.1378 from left to right for R2 group; P = 0.0013, <0.0001, 0.0058, 0.0003, 0.1611, 0.0137, <0.0001, <0.0001, 0.0023, 0.0021 from left to right for R3 group. f, Elisa experiments to determine the expression levels of the wild type and indicated mutants GPBAR in HEK293 cells. Data from three independent measurements are measured as mean ± s.d. Statistical differences between WT and mutations were determined by two-sided one-way ANOVA with Tukey test. n.s., the mutant showed no significantly different the from wild type. (P = 0.1676, 0.3955, 0.0595 from left to right). g, Dose response of the endogenous bile acids and INT-777 induced cAMP accumulation using wild type and mutant GPBAR was monitored by the Glosensor assay. Data from three independent measurements are measured as mean ± SEM.
Extended Data Fig. 6 The active structure of GPBAR in complex with Gs.
a, Structural representation of the important residues participating in GPBAR activation. The toggle switch residues W2466.48 in the A2AR–Gs complex and W2866.48 in β2AR–Gs complex undergoes one helical turn downshift referring to the central TM3 during the transition of the inactive state to active state. Notably, Y2406.51 of GPBAR assumes the same position as W2866.48 in the active β2AR-Gs complex, thus representing the active state. Structures involved in this panel include: inactive A2A (PDB ID 3EML), active A2A-Gs complex (PDB ID 5G53), inactive β2AR (PDB ID 3NYA), active β2AR-Gs complex (PDB ID 3SN6). b, Sequence alignment of the toggle switch W237 and the proline kink P176 in A2AR, β2AR and GPBAR from different species. P176 are colored in red and highlighted in yellow, W237 are colored in green and highlighted in yellow. c, Dose response curve of the INT-777 and other bile acid induced cAMP accumulation in Glosensor assay in cells overexpressing wild type or mutant GPBAR. Data from three independent measurements are measured as mean ± SEM. d, Lack of the compact structural P5.50I3.40F6.44 motif in GPBAR structure. Left, structural rearrangement of the PIF motif during β2AR activation. Right, separation of P5.50L3.40F6.44 in the GPBAR structure. Instead, W2376.48 forms hydrophobic interactions with L1003.40 and V1785.52 to constitute a VLW motif in GPBAR.
Extended Data Fig. 7 Interactions of the α5 helix of Gs with the GPBAR transmembrane core.
a, Interactions of the α5-helix of Gs with the GPBAR transmembrane core. The interface contains more hydrophilic interactions, and the α5 helix is tilted towards TM6 compared to other class A GPCR–Gs complex structures. The eleven hydrophilic interactions include hydrogen bonds formed between the R385, H387 Y391, L393, L394 of the Gs and the E1093.49, Q1955.69, R2216.32 and Q2226.33 of GPBAR; the electrostatic interactions of Gs R385 and E392 with GPBAR D1985.72 and R2216.32, R2246.35 and R286H8 respectively, and finally, a salt bridge formed by the carboxyl end of the α5 helix of Gs with R2216.32 of GPBAR. A comparison of the structure for the interaction of the α5 helix of Gs with the β2AR transmembrane core is shown in Supplementary Fig. 5. b, Comparison of electrostatic potential of transmembrane core in the GPBAR–Gs complex (left panel) and the β2AR–Gs complex (right panel). Colours: red: negative charge; blue: positive charge; white: neutral. c, Comparison of residues contacting the α5 helix of Gs in the GPBAR–Gs complex and the β2AR–Gs complex. Only residues making contacts are shown. Blue- or red-shaded residues are those that make hydrophilic interactions. Ballesteros-Weinstein numbers are given for the TM residues of GPBAR and β2AR. d, Effects of mutations in the GPBAR transmembrane core in the Gs interface on P395–induced cAMP accumulation. The EC50 and Emax data are referred to Supplementary Fig. 5. Data from three independent measurements are measured as mean ± SEM. e, Sequence alignment of different G protein subtypes. Residues responsible for GPBAR ICL3 coupling in Gs and equivalent G protein residues are highlighted in yellow. Notably, Gq/11 carry a Val at the same position equivalent to Gs L394 and an uncharged Asn at the equivalent position to Gs E392, thus reducing their interactions with residues from TM5, 6 and H8 of GPBAR; these divergences may partly contribute to the selective coupling with Gs in preference to Gq by GPBAR.
Extended Data Fig. 8 Interactions of GPBAR ICL3 with Gs.
a, P395–induced Gs (left), Gq (middle) and Gi (right) activity examined by cAMP (GloSensor), Gq-Gγ dissociation or Gi-Gγ dissociation assays, respectively. Data from at least three independent measurements are measured as mean ± SEM. b, Conformational differences of the Gs at GPBAR ICL3-interacting i3 loop, the GTP binding P361–V367 segment and the N292–D295 segment between the GPBAR–Gs complex (yellow orange) and the β2AR–Gs (grey) complex. c, Effects of different ICL3 mutations or ICL3 change of GPBAR on P395-induced cAMP accumulation. ICL3 change: substitution of GPBAR (R201-L214) by the (D234-C265) in β2AR. The EC50 and Emax data was referred to Supplementary Fig. 5. Data from three independent measurements are measured as mean ± SEM. d, Effects of ICL3 mutations in the R/KψXR/KXψXR motifs of V2R (R243A/R247A/R249A/R251A), PF2R (R238A/R241A/HR243A) and EP2 (R242A/R249A) on their agonist-induced cAMP accumulation. Data are shown as mean ± SEM from three independent measurements. e, A cartoon model illustrating the structural differences of the activation and Gs coupling of GPBAR compared to the other class A GPCR–Gs or GPCR–Gi complexes. From the left to right is the inactive GPCR structural model (using β2AR as an example, PDB ID 3NYA), the general GPCR–Gs complex (using β2AR as an example, PDB ID 3SN6), the GPBAR–Gs complex and the NTSR–Gi complex (PDB ID 6OS9). Compared to other class A GPCR–Gs complexes or NTSR–Gi complex, the GPBAR–Gs complex exhibits distinct features, first a larger separation at the TM3-TM6 helices in the centre of receptor region, second the H5 of Gs in GPBAR–Gs complex showing one helical turn downshifting probably due to the direct interaction of the ICL3 of GPBAR with the Gs.
Extended Data Fig. 9 The second ligand-binding pocket of GPBAR.
a, Cryo-EM density of the annular lipid molecules outside of the seven transmembrane bundle in INT-777–GPBAR, P395–GPBAR and LA-PTH–PT1R (PDB, 6NBF) structure respectively. b, Comparison of the lipid binding pocket of GPBAR (green) with that of the P2Y1R (tan; PDB, 4XNV) and GPR40 (light bule; PDB, 5TZY). A similar lipid binding site for GPCR P2Y1 (PDB ID 4XNV) and an allosteric modulator site for GPR40 have been reported. c, Fitting of the cholesterol, CHS into the electron density of the second ligand binding site in the P395-GPBAR–Gs complex cryo-EM structure. The cholesterol can be easily fit into the electron density, whereas the CHS cannot be fit into the electron density. The cholesterol may come from the plasma membrane. d, Fitting of the INT-777, cholesterol into the electron density of the second ligand binding site in the INT-777–GPBAR–Gs complex cryo-EM structure. The INT-777 and cholesterol can be fit into the electron density. Computational simulation indicated that both the GPBAR and the INT-777 bound at orthosteric site exhibit least r.m.s.d. fluctuations in the presence of the INT-777, but not the cholesterol, CHS or no ligand at this lipid binding site (e). e, Molecular dynamics simulation of INT-777–GPBAR–CHS, INT-777–GPBAR–Cholesterol, INT-777–GPBAR–INT-777 and INT-777–GPBAR trajectories. (Top) RMSDs of allosteric ligands CHS (blue), choslesterol (green) or INT-777 (red) of 200 ns MD simulation. (Middle) RMSDs of orthosteric INT-777 of four structures, trajectory of INT-777-GPBAR is colored in purple. (Bottom) RMSDs of the backbone of GPBAR of four structures. Values were calculated based on the initial complex state after equilibration (0 ns). f, The effects of mutations of residues in second ligand binding pocket on the allosteric effects of different bile acids. The original data was referred to Supplementary Table 9. Values are the mean ± SEM of three independent experiments for the wild type (WT) and mutants. Statistical differences between WT and mutations were determined by two-sided one-way ANOVA with Tukey test. *P < 0.05, **P < 0.01,***P < 0.001, n.s., no significant difference; for CA group: P = 0.196, < 0.0001, 0.0088, 0.0734, 0.0538, 0.0020, 0.0011, 0.0005 (left to right); for DCA group: P = 0.173, < 0.0001, < 0.0001, 0.1009, 0.4789, 0.0001, 0.0009, < 0.0001 (left to right); for GCA group: P = 0.0529, 0.0006, 0.0002, 0.6314, 0.4776, < 0.0001, 0.0136, 0.0019 (left to right); for TCA group: P = 0.5217, < 0.0001, 0.0180, 0.2203, 0.0651, < 0.0001, 0.0004, < 0.0001 (left to right); for TDCA group: P = 0.7280, 0.0003, 0.0007, 0.7168, 0.3524, 0.0003, 0.0001, 0.0001 (left to right).
Extended Data Fig. 10 Effect of the alanine scanning mutagenesis of the potential residues involved in biased property of GPBAR.
a, G protein or β-arrestin signalling properties of GPBAR agonists. HEK293 cells were transfected with cDNAs encoding GPBAR and stimulated with various bile acids or bile acid derivatives. The cAMP level was measured using a GloSensor assay, whereas a bioluminescence resonance energy transfer (BRET) assay was used to measure β-arrestin-2 recruitment. The EC50 and Emax values are the mean ± s.d. of at least 3 independent experiments. ND, not determined due to low signal. b, EM density of the ICL1, ECL1, ECL2 and ECL3 in INT-777-GPBAR-Gs complex and P395-GPBAR-Gs complex. c, Plot of the distance root-mean-square deviations (RMSDs) of each residue between INT-777–GPBAR and P395–GPBAR structures. The horizontal and vertical axes indicate the amino acid sequence of the GPBAR and the RMSDs (Cα deviations) for every residue, respectively. The red, blue, green and grey dots represent Cα deviations that range from >3, 2~3, 1~2 or <1, respectively. d, Dose response of the P395 induced β-arrestin-2 recruitment was examined by BRET assay and the dose response of the P395 induced cAMP accumulation was monitored by the Glosensor assay using wild type and mutant GPBAR, respectively. Data from three independent measurements are measured as mean ± SEM. e, Elisa experiments to assay the expression levels of the wild type and indicated mutants of GPBAR in HEK293 cells. Data from three independent experiments are presented as mean ± s.d. Statistical differences between WT and mutations were determined by two-sided one-way ANOVA with Tukey test. n.s., the mutant showed no significant difference between wild type. and mutants (P = 0.3521, 0.6531, 0.5764, 0.6377 from left to right).
Supplementary information
Supplementary information
This file contains Supplementary Figures 1-5 and Supplementary Tables 1-9
Rights and permissions
About this article
Cite this article
Yang, F., Mao, C., Guo, L. et al. Structural basis of GPBAR activation and bile acid recognition. Nature 587, 499–504 (2020). https://doi.org/10.1038/s41586-020-2569-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2569-1
This article is cited by
-
Mogrol stimulates G-protein-coupled bile acid receptor 1 (GPBAR1/TGR5) and insulin secretion from pancreatic β-cells and alleviates hyperglycemia in mice
Scientific Reports (2024)
-
Bitter taste receptor activation by cholesterol and an intracellular tastant
Nature (2024)
-
G protein-coupled receptors (GPCRs): advances in structures, mechanisms, and drug discovery
Signal Transduction and Targeted Therapy (2024)
-
Structure of GPR101–Gs enables identification of ligands with rejuvenating potential
Nature Chemical Biology (2024)
-
Ursodeoxycholic acid induces sarcopenia associated with decreased protein synthesis and autophagic flux
Biological Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.