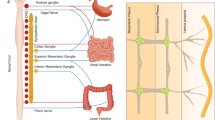
Abstract
Connections between the gut and brain monitor the intestinal tissue and its microbial and dietary content1, regulating both physiological intestinal functions such as nutrient absorption and motility2,3, and brain-wired feeding behaviour2. It is therefore plausible that circuits exist to detect gut microorganisms and relay this information to areas of the central nervous system that, in turn, regulate gut physiology4. Here we characterize the influence of the microbiota on enteric-associated neurons by combining gnotobiotic mouse models with transcriptomics, circuit-tracing methods and functional manipulations. We find that the gut microbiome modulates gut-extrinsic sympathetic neurons: microbiota depletion leads to increased expression of the neuronal transcription factor cFos, and colonization of germ-free mice with bacteria that produce short-chain fatty acids suppresses cFos expression in the gut sympathetic ganglia. Chemogenetic manipulations, translational profiling and anterograde tracing identify a subset of distal intestine-projecting vagal neurons that are positioned to have an afferent role in microbiota-mediated modulation of gut sympathetic neurons. Retrograde polysynaptic neuronal tracing from the intestinal wall identifies brainstem sensory nuclei that are activated during microbial depletion, as well as efferent sympathetic premotor glutamatergic neurons that regulate gastrointestinal transit. These results reveal microbiota-dependent control of gut-extrinsic sympathetic activation through a gut–brain circuit.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
TRAP RNA sequencing and 16S sequencing data that support the findings of this study have been deposited in the Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/) under accession code GSE145986. Raw data are provided for all figures with bar graphs, namely Figs. 1–4 and Extended Data Figs. 1–10. Source data are provided with this paper.
Change history
20 August 2020
A Correction to this paper has been published: https://doi.org/10.1038/s41586-020-2657-2
References
Furness, J. B., Rivera, L. R., Cho, H. J., Bravo, D. M. & Callaghan, B. The gut as a sensory organ. Nat. Rev. Gastroenterol. Hepatol. 10, 729–740 (2013).
Han, W. et al. A neural circuit for gut-induced reward. Cell 175, 665–678 (2018).
Williams, E. K. et al. Sensory neurons that detect stretch and nutrients in the digestive system. Cell 166, 209–221 (2016).
Fung, T. C., Olson, C. A. & Hsiao, E. Y. Interactions between the microbiota, immune and nervous systems in health and disease. Nat. Neurosci. 20, 145–155 (2017).
Veiga-Fernandes, H. & Mucida, D. Neuro-immune interactions at barrier surfaces. Cell 165, 801–811 (2016).
Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P. & Heintz, N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protocols 9, 1282–1291 (2014).
Mei, Q., Mundinger, T. O., Kung, D., Baskin, D. G. & Taborsky, G. J., Jr. Fos expression in rat celiac ganglion: an index of the activation of postganglionic sympathetic nerves. Am. J. Physiol. Endocrinol. Metab. 281, E655–E664 (2001).
Biggs, M. B. et al. Systems-level metabolism of the altered Schaedler flora, a complete gut microbiota. ISME J. 11, 426–438 (2017).
Atarashi, K. et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500, 232–236 (2013).
Gabanyi, I. et al. Neuro-immune interactions drive tissue programming in intestinal macrophages. Cell 164, 378–391 (2016).
Braniste, V. et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci. Transl. Med. 6, 263ra158 (2014).
Rivera-Chávez, F. et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19, 443–454 (2016).
Koh, A., De Vadder, F., Kovatcheva-Datchary, P. & Bäckhed, F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell 165, 1332–1345 (2016).
Nøhr, M. K. et al. Expression of the short chain fatty acid receptor GPR41/FFAR3 in autonomic and somatic sensory ganglia. Neuroscience 290, 126–137 (2015).
Selwyn, F. P., Csanaky, I. L., Zhang, Y. & Klaassen, C. D. Importance of large intestine in regulating bile acids and glucagon-like peptide-1 in germ-free mice. Drug Metab. Dispos. 43, 1544–1556 (2015).
Harach, T. et al. TGR5 potentiates GLP-1 secretion in response to anionic exchange resins. Sci. Rep. 2, 430 (2012).
Young, J. B. & Landsberg, L. Suppression of sympathetic nervous system during fasting. Science 196, 1473–1475 (1977).
Chi, J. et al. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metabolism 27, 226–236 (2018).
Browning, K. N. & Travagli, R. A. Central nervous system control of gastrointestinal motility and secretion and modulation of gastrointestinal functions. Compr. Physiol. 4, 1339–1368 (2014).
Barbara, G. et al. Interactions between commensal bacteria and gut sensorimotor function in health and disease. Am. J. Gastroenterol. 100, 2560–2568 (2005).
Allen, W. E. et al. Thirst-associated preoptic neurons encode an aversive motivational drive. Science 357, 1149–1155 (2017).
Renier, N. et al. Mapping of brain activity by automated volume analysis of immediate early genes. Cell 165, 1789–1802 (2016).
Fung, C. et al. Cholinergic submucosal neurons display increased excitability following in vivo cholera toxin exposure in mouse ileum. Front. Physiol. 9, 260 (2018).
Young, A. A. Brainstem sensing of meal-related signals in energy homeostasis. Neuropharmacology 63, 31–45 (2012).
Pavlov, V. A., Chavan, S. S. & Tracey, K. J. Molecular and functional neuroscience in immunity. Annu. Rev. Immunol. 36, 783–812 (2018).
Zeng, W. Z. et al. PIEZOs mediate neuronal sensing of blood pressure and the baroreceptor reflex. Science 362, 464–467 (2018).
Berthoud, H. R., Jedrzejewska, A. & Powley, T. L. Simultaneous labeling of vagal innervation of the gut and afferent projections from the visceral forebrain with dil injected into the dorsal vagal complex in the rat. J. Comp. Neurol. 301, 65–79 (1990).
Wang, F. B. & Powley, T. L. Topographic inventories of vagal afferents in gastrointestinal muscle. J. Comp. Neurol. 421, 302–324 (2000).
Moriyama, S. et al. β2-adrenergic receptor-mediated negative regulation of group 2 innate lymphoid cell responses. Science 359, 1056–1061 (2018).
Matheis, F. et al. Adrenergic signaling in muscularis macrophages limits infection-induced neuronal loss. Cell 180, 64–78 (2020).
Zhang, B. et al. Hyperactivation of sympathetic nerves drives depletion of melanocyte stem cells. Nature 577, 676–681 (2020).
Lai, N. Y. et al. Gut-innervating nociceptor neurons regulate Peyer’s patch microfold cells and SFB levels to mediate Salmonella host defense. Cell 180, 33–49 (2020).
Yadav, V. K. et al. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 135, 825–837 (2008).
Gomes-Neto, J. C. et al. A real-time PCR assay for accurate quantification of the individual members of the Altered Schaedler Flora microbiota in gnotobiotic mice. J. Microbiol. Methods 135, 52–62 (2017).
Bai, L. et al. Genetic identification of vagal sensory neurons that control feeding. Cell 179, 1129–1143 (2019).
Ye, L. et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell 165, 1776–1788 (2016).
Kennedy, H. S., Jones, C., III & Caplazi, P. Comparison of standard laminectomy with an optimized ejection method for the removal of spinal cords from rats and mice. J. Histotechnol. 36, 86–91 (2013).
Bray, N. L., Pimentel, H., Melsted, P. & Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016); erratum 34, 888 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Ertürk, A. et al. Three-dimensional imaging of solvent-cleared organs using 3DISCO. Nat. Protocols 7, 1983–1995 (2012).
Abe, C. et al. C1 neurons mediate a stress-induced anti-inflammatory reflex in mice. Nat. Neurosci. 20, 700–707 (2017).
Haak, B. W. et al. Impact of gut colonization with butyrate-producing microbiota on respiratory viral infection following allo-HCT. Blood 131, 2978–2986 (2018).
Acknowledgements
We thank all members of the Mucida laboratory, past and present, for assistance with experiments, fruitful discussions and critical reading of the manuscript. In particular we thank: A. Rogoz for maintaining gnotobiotic mice; S. Gonzalez for maintaining SPF mice; T. Rendon and B. Lopez for genotyping; C. Nowosad for setting up the ASF; A. North, C. Pyrgaki, T. Tong, C. Rico and P. Ariel of The Rockefeller University Bio-imaging Research Center for assistance with light-sheet microscopy and image analysis; The Rockefeller University Genomics Center for RNA sequencing; Rockefeller University employees for continuous assistance; T. Y. Oliveira for valuable discussions on TRAP-seq approaches and help with initial analysis; S. Poliak (Kallyope) for help in setting up tracing experiments and valuable input throughout the project; K. Loh and A. Ilanges for fruitful experimental discussions; J. Friedman and V. Ruta for the use of laboratory equipment; A. Lockhart, V. Jové and V. Ruta for critical reading of the manuscript; V. Jové for critical reading of reviewer responses; L. Vosshall, M. Nussenzweig, G. Victora and J. Lafaille and their respective laboratory members for fruitful discussions and suggestions; D. Littman and M. Xu for providing faeces from SFB monocolonized mice; D. Artis and G. Sonnenberg for providing germ-free mice and Clostridium consortium colonized mice; N. Arpaia for Gpr43−/− mice; S. Mehandru for Gpr109a−/− Gpr43−/− mice; J. Pluznick, J. Gordon and M. Yanagisawa for Gpr41−/− mice; I. Chiu and J. Wood for Nav1.8Cre mice; C. Woolf and R. Kuhner for SNSCre mice; A. Ramer-Tait for ASF caeca; J. Faith for germ-free mice; G. Donaldson for A. muciniphilia and B. fragilis monocolonized mice; and C. Nowosad for OligoMM12 mice. We finally thank R. U. Muller for his inspiration. This work was supported by National Institute of Diabetes and Digestive Kidney Diseases (NIDDK) grant K99 DK120869 (to M.S.); NIH Virus Center grant P40 OD010996; a NIH F31 Kirchstein Fellowship (to P.A.M.); National Center for Advancing Translational Sciences (NCATS) NIH grant UL1TR001866 (to P.A.M. and D.M.); a Philip M. Levine Fellowship (to P.A.M.); a Kavli Graduate Fellowship (to P.A.M.); a Kavli Postdoctoral Fellowship (to M.S.); an Anderson Graduate Fellowship (to F.M.); a Leon Levy Fellowship in Neuroscience (to J.d.M.); the Leona M. and Harry B. Helmsley Charitable Trust (to D.M.); the Kenneth Rainin Foundation (to D.M.); a Burroughs Wellcome Fund PATH Award (to D.M.); and Transformative grant R01DK116646 (to D.M).
Author information
Authors and Affiliations
Contributions
P.A.M. initiated, designed, performed and analysed the research, helped to supervise the research, and wrote the manuscript. M.S. performed brain AdipoClear and ClearMap analysis, provided guidance on brain experiments, analysed data and helped with manuscript preparation. F.M. performed experiments, analysed data,and helped with figure and manuscript preparation. P.W. performed stereotaxic brain injections, provided technical advice and reviewed the manuscript. Z.K. performed experiments, analysed research and reviewed the manuscript. A.I. performed stereotaxic brain injections of the NTS and area postrema, provided technical advice, and reviewed the manuscript. K.P. performed i.c.v. and stereotaxic viral injection experiments. T.B.R.C. gave guidance on bioinformatic approaches, wrote analysis script, and performed 16S sequencing analysis. I.d.A, W.H. and M.P. gave expert advice and technical support with vagal nerve experiments, including nerve recordings. K.H. and M.F. provided the Clostridium consortium and performed faecal SFB SCFA measurements. A.R. extracted, ran and validated the quantification SCFA quantifications. A.J.P. developed, validated and analysed the SCFA quantifications. J.R.C. supervised and analysed SCFA quantifications. J.d.M. performed electrophysiology experiments and analysed the data. D.M. initiated, designed and supervised the research, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Peer review information Nature thanks Ulrika Marklund, Scott Russo and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Comparison of extrinsic EAN organization.
a, Number of CTB+ neurons per left (L) and right (R) (n = 5 ganglia from independent mice) nodose ganglion (NG) of C57BL/6J SPF mice injected with CTB555 into the caecum. b, Number of CTB+ neurons per CG-SMG of mice injected with CTB555 into the caecum (n = 4 ganglia from independent mice). c, Retrograde CTB tracing from intestinal segments to DRG at thoracic level (Th) 10. Images representative of duodenum (n = 3 averaged pairs of ganglia from independent mice), ileum (n = 3 averaged pairs of ganglia from independent mice) and proximal colon (n = 3 averaged pairs of ganglia from independent mice). d, Left, scheme showing retrograde tracing to the DRG. Right, distribution of labelled neurons in the DRG of mice injected with CTB555 in the duodenum (n = 3 averaged pairs of ganglia from independent mice), ileum (n = 3 averaged pairs of ganglia from independent mice), proximal colon (n = 3 averaged pairs of ganglia from independent mice) and mid-distal colon (n = 2 averaged pairs of ganglia from independent mice). L, lumbar. e, Left, number of CTB+ neurons and right, representative images of individual superior cervical (SCG) (n = 10 ganglia from 7 mice) and stellate (n = 8 ganglia from 4 mice) ganglia of mice injected with CTB647 in the ileum and proximal colon. f, Loop of distal ileum from sham-operated and locally denervated (DNx) mice. Representative of n = 7 mice. g, Representative right NG (RNG) of mice injected with CTB594 into the distal ileum of sham-operated (n = 7 ganglia) or DNx mice (n = 6 ganglia). h, Number of CTB+ neurons in the CG-SMG and NG of sham-operated (L-NG, n = 5 ganglia; R-NG, n = 7 ganglia; CG-SMG, n = 5 ganglia) or DNx mice (L-NG, n = 5 ganglia; R-NG, n = 7 ganglia; CG-SMG, n = 4 ganglia) injected with CTB594/647 in the distal ileum. i, Images of NG from mice injected with CTB488 into the jejunum (n = 2 mice). j, Anatomical verification of subdiaphragmatic vagotomy (sdVx), showing enlarged stomachs (top) and successful severing of the vagal nerves below the diaphragm (bottom). Images representative of n = 10 mice. k, Immunofluorescence images of NG from sham-operated or sdVx mice three days after i.p. FluoroGold injection (native fluorescence). Images representative of n = 5 ganglia. l, Immunofluorescence images of NG from sham-operated or sdVx mice injected with CTB488 into the stomach. Images representative of n = 5 ganglia. m, Immunofluorescence images of NG from sdVx mice injected with CTB488 in the ileum/proximal colon or from sham-operated mice injected with CTB488 in the proximal colon. Images representative of n = 2 ganglia. n, Dual CTB tracing in DRG Th9 from the duodenum (green arrows) and ileum (red arrows). Representative of n = 3 mice. o–q, Dual CTB tracing in the CG-SMG and NG from the: o, duodenum (green arrows) and ileum (red arrows); p, spleen (green arrows) and distal colon (red arrows); q, duodenum and stomach (white arrows indicate dual-labelled neurons). Images representative of n = 3 ganglia from independent mice. c, e, g, i, k–q, Scale bars, 50 μm. a, b, d, e, h, Means ± s.d. a, h, Two-tailed unpaired t-test. g, i, k, l, Dashed lines highlight the NG. m–q, Arrows indicate CTB+ cells.
Extended Data Fig. 2 Microbial depletion leads to changes in extrinsic EAN.
a, Immunofluorescence images of the nodose ganglion (NG), CG-SMG and DRG from Snap25RiboTag SPF mice using anti-haemagglutinin antibody (HA, green) and NeuroTrace (red). Images representative of n = 4. b, Immunofluorescence images of the NG, CG-SMG and DRG from Snap25RiboTag mice germ-free mice using anti-HA antibody (green) and NeuroTrace (red). Images representative of n = 2. c–e, Volcano plots of differentially expressed genes by: c, NG and DRG Th9; d, DRG Th9 and CG-SMG; and e, NG and CG-SMG of Snap25RiboTag SPF mice. f–h, Immunofluorescence images of CG-SMG from: f, C57BL/6J SPF mice using anti-TH (green) and anti-NPY (red) antibodies; g, ChattdTomato mice using anti-cFos antibody (red) and native tdTomato fluorescence (green); and h, C57BL/6J SPF mice using anti-TH (green) and anti-SST (red) antibodies. Images representative of n = 3 ganglia. i, Volcano plot showing differentially expressed genes by DRG Th9 from germ-free and SPF Snap25RiboTag mice. Red/blue dots represent a log2-transformed fold change of more than 0.5; italicized parentheses represent a log2-transformed fold change of more than 1. j, Immunofluorescence images of the CG-SMG from C57BL/6J SPF mice three days after i.p. FluoroGold injection (native fluorescence). The dotted line outlines the FluoroGold+ signal. Image representative of n = 4 ganglia. k, Immunofluorescence images of CG-SMG from acutely stressed C57BL/6J SPF mice three days after i.p. FluoroGold injection using with anti-cFos antibody (red) and native FluoroGold fluorescence. Image representative of n = 4 ganglia. l, Immunofluorescence images of CG-SMG from acutely stressed C57BL/6J SPF mice stained for cFos (white). White arrows indicate neuronal cFos+ nuclei; red arrows indicate non-neuronal cFos+ nuclei. Scale bar, 10 μm. Image representative of n = 4 ganglia. m, n, Most abundant bacterial genera (m) and Shannon diversity index (n) in 16S sequencing of caecal samples from C57BLl6/J mice 24 h after PBS treatment (n = 5) or streptomycin (strep) treatment (n = 5), as determined by the phyloseq R package. a, b, f–h, j, k, l, Scale bars, 50 μm. g, Arrows indicate cFos+ cells and stars indicate tdTomato+ cells. c–e, i, Numbers (n) of independent biological samples analysed are indicated in parentheses. c–e, Red/blue dots represent a log2-transformed fold change of more than 3. c–e, i, ‘padj’ denotes two-tailed Benjamini–Hochberg test P value computed from two-sided Wald test P values less than 0.05.
Extended Data Fig. 3 Effects of modulation of SCFAs on gut sympathetic neurons.
a–f, Quantification of SCFAs (acetate, propionate and butyrate) in the caecal (a–d, f) or faecal (e) contents of: a, VAMN- (n = 5) and Splenda- (n = 5) treated C57BL/6J mice; b, PBS- (n = 5) and streptomycin- (n = 5) treated C57BL/6J mice; c, single antibiotic (n = 5 for each antibiotic) and Splenda- (n = 5) treated C57BL/6J mice; d, C57BL/6J GF mice (n = 3) colonized with ASF (n = 2) or Clostridium consortium (n = 4); e, C57BL/6J germ-free (GF) mice (n = 5), GF mice monocolonized with SFB (n = 5), or SPF (n = 5) mice; and f, GF mice (n = 5), SPF mice (n = 5), and GF mice colonized with A. muciniphilia (n = 3), B. fragilis (n = 3), or OligoMM12 (n = 4). g, Caecal weight of SPF (n = 6) and OligoMM12 (n = 14) mice. h, Images of dissected caecum (in six-well cell culture plate) from mice treated with Splenda, ampicillin, metronidazole, vancomycin or neomycin for two weeks. Each representative of n = 5 caeca from separate mice. i, Number of cFos+ neurons in the CG-SMG of C57BL/6J mice treated with methylcellulose (n = 5) or mannitol (n = 5). j, Number of Salmonella CFUs per milligram of faeces from Salmonella-infected CBA/J mice (n = 5), compared with luria broth (LB)-gavaged control mice (n = 5). k, Number of cFos+ neurons in the CG-SMG of CBA/J mice orally inoculated with Salmonella (n = 10) or given oral gavage of LB (n = 10). The dashed line indicates the average number of cFos+ neurons for antibiotic-treated mice (320). l, Quantification of SCFAs in the caecal contents of CBA/J mice orally inoculated with Salmonella (n = 4) or given oral gavage of LB (n = 5). m–o, Number of cFos+ neurons in the CG-SMG of: m, C57BL/6J SPF mice 24 h post oral gavage with streptomycin and treated with SCFA-supplemented or sodium (Na)-containing water; n, C57BL/6J SPF mice treated with intracerebroventricular (i.c.v.) artificial cerebrospinal fluid (aCSF) (n = 5) or SCFAs in aCSF (n = 4) post streptomycin treatment; o, C57BL/6J GF mice 24 h post treatment with tributyrin (n = 5) or PBS (n = 5); p, C57BL/6J SPF mice treated with tributyrin (n = 7) or PBS (n = 6) 24 h post oral gavage of streptomycin; and q, CBA/J SPF mice treated with tributyrin (n = 5) or PBS (n = 5) 24 h post oral gavage of streptomycin. r, Quantification of SCFAs in caecal samples of GF mice given consecutive oral gavage of PBS (‘GF’; n = 2) or tributyrin (‘GF + tributyrin’; n = 3). s, Caecal weight of mice given oral gavage of streptomycin (strep) followed by treatment with PBS (n = 5) or tributyrin (n = 5). t–v, Number of cFos+ neurons in the CG-SMG of: t, Gpr109a–/– (n = 3) and littermate control Gpr109a+/– mice (n = 3); u, Gpr109a–/–Gpr43–/– mice (n = 8); and v, Gpr41–/– and littermate control Gpr41+/+ mice. a–g, i–t Means ± s.d. a, b, d, g, i, k–q, s–v, Two-tailed unpaired t-test. c, e, f, One-way ANOVA with Tukey’s multiple comparisons. a–d, f, l, The dashed line indicates the lowest limit of detection. t–v, The dashed line indicates the average number of cFos+ neurons/CG-SMG for GF mice (334).
Extended Data Fig. 4 Effects of microorganism-modulated host factors on sympathetic activation and gut motility.
a, Number of cFos+ neurons in the CG-SMG of C57BL/6J mice treated with cholestyramine after oral gavage of PBS (n = 5) or streptomycin (n = 5). b, Number of cFos+ neurons in the CG-SMG of C57BL/6J mice with bile-duct cauterization after oral gavage of PBS (n = 3) or streptomycin (n = 4). c, Agarose gel image from Tph1 recombination PCR analysis of colon samples. The lower bands correspond to the recombined allele in tamoxifen-treated VilΔTph1 (n = 3) mice, compared with VilCreERT2− (n = 3) control mice. d, Serotonin (5-HT) concentration in colonic tissues from VilΔTph (n = 5), VilTph1fl/+ (n = 7) and VilCreERT2− (n = 7) mice two weeks post tamoxifen injection. e, f, Gastrointestinal transit time (e) and number of cFos+ neurons (f) in the CG-SMG of VilΔTph (n = 5) or control mice (n = 8) two weeks post tamoxifen injection. g–i, Number of cFos+ neurons in the CG-SMG (g), colonic motility (h) and gastrointestinal transit time (i) of C57BL/6J mice 4 h post i.p. injection of Exendin-4 (n = 5) or saline (n = 5). j, Gastrointestinal transit time of C57BL/6J mice 5 min post i.p. injection of Exendin-4 with saline (n = 10) or guanethidine (n = 9). k, Number of cFos+ neurons in the CG-SMG of C57BL/6J mice 4 h post i.p. injection of saline (n = 5) or Exendin-9 (n = 5) and 24 h post streptomycin treatment. l, Number of cFos+ neurons in the CG-SMG of Glp1r–/– mice treated for two weeks with broad-spectrum antibiotics (VAMN, n = 5) or Splenda (n = 4) in drinking water. m, n, Number of cFos+ neurons in the CG-SMG of C57BL/6J mice either: m, 4 h post i.p. injection of saline (n = 5) or PYY (50 mg kg−1) (n = 5); or n, 4 h post i.p. injection of saline (n = 5) or PYY (50 mg kg−1) (n = 5) in mice treated with streptomycin 24 h previously. a, b, d–n, Mean ± s.d. a, b, e–n, Two-tailed unpaired t-test. d, One-way ANOVA with Tukey’s multiple comparisons. e, h–j, The dashed lines indicates the maximum time allowed per mouse for each motility measurement. l, The dashed line indicates the average number of cFos+ neurons per CG-SMG for GF mice (334). f, g, m, The dashed line indicates the average number of cFos+ neurons per CG-SMG for antibiotic-treated mice (320).
Extended Data Fig. 5 CG-SMG activation is unlikely to require direct sensing of microorganisms or viscerofugal input.
a–c, Injection of AAVrg–CAG–FLEX–-tdTomato into the CG-SMG of Snap25Cre mice for tracing of fibres in the intestine. a, Left, scheme and right, immunofluorescence images of the CG-SMG using anti-TH antibody (green) and native tdTomato fluorescence (red). Image representative of n = 2. b, Light-sheet three-dimensional (3D) reconstruction and optical section of the colon (left) or caecum (right). Left-hand images, scale bars, 300 μm (left) and 200 μm (right). Right-hand images, scale bars, 500 μm (left) and 200 μm (right). Images representative of n = 3. c, Immunofluorescence images of the ileum (top) and colon myenteric plexus (bottom) using anti-TH antibody (green) and native tdTomato fluorescence (red). Image representative of n = 2. d–f, AAVrg–hSyn1–Cre injection into the CG-SMG of Rosa26lsl-tdTomato mice for tracing of fibres in the intestine. d, Left, scheme and right, immunofluorescence image of the CG-SMG using native Tomato fluorescence (red). The dashed line indicates the borders of the CG-SMG. Image representative of n = 2. e, Immunofluorescence image of the ileum myenteric plexus using native Tomato fluorescence (red). Image representative of n = 2. f, Immunofluorescence image of the ileum villi using anti-TH(green) antibody and native tdTomato fluorescence (red). Image representative of n = 2. g, Immunofluorescence stitched images of the colon myenteric plexus from C57BL/6J mice treated with 6-OHDA using anti-TH antibody (red). Dashed lines indicate the mesenteric border. Scale bar, 500 μm. Image representative of n = 10. h, Number of cFos+ neurons in the CG-SMG of C57BL/6J mice treated with 6-OHDA before PBS (n = 5) or streptomycin (n = 5) treatment. i, Transcripts per million (TPM) as calculated by Kallisto alignment for Ffar3 expression by CG-SMG neurons isolated from SPF (n = 3) and GF (n = 3) Snap25RiboTag mice (TRAP-seq analyses). j, Immunofluorescence image, using anti-cFos antibody, of the CG-SMG from C57BL/6J mice i.p. injected with tributyrin. Image representative of n = 4. k, Left, scheme representing injection of CT555 into the CG-SMG for visualization of viscerofugal neurons. Right, immunofluorescence image of the ileum myenteric plexus using native CTB555 fluorescence. White arrows indicate CTB+ neurons. Image representative of n = 3. l, Left, scheme representing injection of AAV6–CAG–FLEX–tdTomato into the intestine of Snap25Cre mice for tracing of fibres. Right, immunofluorescence image of CG-SMG showing tdTomato+ fibres originating, in part, from the intestine. The CG-SMG is outlined with dashed lines. Image representative of n = 2. m–t, PBS- and streptomycin-treated C57BL/6J mice 24 h post gavage received retrograde CTB594 injection into the CG-SMG. m–v, Analysis of the ileum (m–o, s, u; n = 5 PBS, n = 5 streptomycin) and colon (p–r, t, v; n = 4 PBS, n = 5 streptomycin). m, p, Total number of ANNA-1+ neurons; n, q, percentage of cFos+ neurons within ANNA-1+ neurons; o, r, percentage of cFos+ and CTB+ neurons within ANNA-1+ neurons. s, t, Immunofluorescence staining of myenteric plexus neurons from the ileum (s) or colon (t) using anti-ANNA-1 (green) and anti-cFos (white) antibodies combined with native CTB594 fluorescence. Arrows indicate CTB+ cFos+ cells. u, v, Percentage of CTB+ neurons among ANNA-1+ neurons in the ileum (u) or colon (v). a, c–f, j–l, s, t, Scale bars, 50 μm. h, i, m–r, u, v, Means ± s.d. h, i, m–r, u, v, Two-tailed unpaired t-test.
Extended Data Fig. 6 Mapping and characterization of sympathetic preganglionic and premotor populations that connect to the intestine.
a–c, Number of cFos+ neurons in the CG-SMG of: a, Splenda-treated fasted (n = 5) or fed (n = 6) or VAMN-treated fasted (n = 4) or fed (n = 4); b, streptomycin-treated fasted (n = 5) or fed (n = 5); and c, GF fasted (n = 6) or fed (n = 5) C57BL/6J mice. d, Caecal weights of Splenda-treated fasted (n = 5) or fed (n = 6), VAMN-treated fasted (n = 5) or fed (n = 4), streptomycin-treated fasted (n = 8) or fed (n = 7) and GF fasted (n = 6) or fed (n = 5) C57BL/6J mice. e, Optical projection of 3DISCO-cleared spinal cord, showing sympathetic preganglionic CTB+ neurons from C57BL/6J SPF mice injected with CTB555 into the CG-SMG. Scale bar, 20 μm. IC, intercalated nucleus; IML, intermediolateral cell column; IPPe, central autonomic area. Image representative of n = 3. f–h, 3D reconstruction of calAdipoClear-cleared CG-SMG (f), spinal cord (sympathetic preganglionic neurons) (g) and intact spinal column (h), from C57BL/6J SPF mice four days post injection of PRV–RFP (red) into the ileum, stained with anti-TH (green) antibody. Scale bars in f, g, 300 μm. Image representative of n = 3. In h, anti-TH antibody is in white and autofluorescence is in green; the dashed outlines identify relevant spinal cord structures. Scale bars, 400 μm. Image representative of n = 3. i, Immunofluorescence images of brainstem slices from VGATL10GFP mice four days post injection of PRV–RFP into the ileum or peritoneum. Dashed lines highlight relevant areas. Images representative of n = 2. j, Immunofluorescence images of the dorsal vagal complex in brainstem slices from C57BL/6J mice subjected to subdiaphragmatic vagotomy (sdVx), 4 days post injection of PRV-RFP in the ileum. Dashed lines highlight relevant areas. CC, central canal. Images representative of n = 2. k, Immunofluorescence image of medullary regions in a brainstem slice from C57BL6/J mice subjected to sdVx, four days post injection of PRV–RFP into the ileum. Dashed lines highlight relevant areas. Images representative of n = 2. l, ClearMap analysis of C57BL6/J mice three and four days post injection of PRV–RFP into the ileum, showing the caudal pontine reticular nucleus. Scale bars, 1 mm. Images representative of n = 3. m, Immunofluorescent images of neuronal subsets within the RPa and Gi in mice four days post PRV–RFP injection into the ileum. Native fluorescence for RFP is red; sections were stained with anti-5-HT (green), anti-VGLUT2 (green) and anti-TH (green) antibodies. Dashed lines highlight the RPa. White arrows indicate VGLUT2+ cells. Images representative of n = 2. n, Immunofluorescent images of the brainstem from mice four days post injection of PRV–GFP (duodenum) and PRV–RFP (ileum). Relevant brainstem regions are highlighted. Representative images of n = 3. i–k, m, n, Scale bars, 50 μm. a–d, Means ± s.d. a, d, One-way ANOVA with Tukey’s multiple comparisons. b, c, Two-tailed unpaired t-test.
Extended Data Fig. 7 Gut-connected sympathetic pre-motor brainstem regions modulate intestinal motility.
a, Immunofluorescence images of brain slices from C57BL/6J SPF and GF mice stained with anti-cFos (black) and anti-TH (red) antibodies. White dashed lines outline RPa, Gi, LPGi and RVLM. Images representative of n = 2. b, Left, percentage of TRAP+ cells among PRV–GFP+ cells, and right, total number of TRAP+ cells in the LPGi/RVLM of FosTRAP2:tdTomato mice. Mice were treated with streptomycin (n = 4) or PBS (n = 4), and 24 h later with 4-OHT. c, Immunofluorescent images of brain slices from VGAT:hM3DqGi/RPa and VGLUT2:hM3DqLPGi/RVLM mice stained with anti-cFos (green) and anti-RFP (red) antibodies. White dashed lines outline relevant areas. Arrows indicate mCherry+ cFos+ cells. Images representative of n = 3. d, Top, colonic motility and bottom, gastrointestinal transit time of C57BL6/J mice treated with 1 mg kg−1 C21 (n = 5) or saline (n = 5). e, Gastrointestinal transit time of VGLUT2:hM3DqLPGi/RVLM (n = 14) or hM3DqLPGi/RVLM (n = 14) mice treated with saline. f, Total faecal output of VGLUT2:hM3DqLPGi/RVLM (n = 14) and hM3DqLPGi/RVLM (n = 10) mice treated with 1 mg kg−1 C21. g, h, Open-field test velocity of: left, VGLUT2:hM3DqLPGi/RVLM (n = 14) and hM3DqLPGi/RVLM (n = 8) mice; and right, VGAT:hM3DqGi/RPa (n = 6) and hM3DqGi/RPa (n = 6) mice, treated with saline (g) or 1 mg kg−1 C21 (h). a, c, Scale bars, 50 μm. b, d–h, Means ± s.d. b, d–h, Two-tailed unpaired t-test. d, e, Dashed lines indicate the maximum time allowed per mouse for motility measurement.
Extended Data Fig. 8 Mapping and characterization of the dorsal vagal complex connected to distal intestine.
a, ClearMap analysis of the nucleus tractus solitarius (NTS)/dorsal motor nucleus of the vagus nerve (DMV) on days 3 and 4 post PRV–RFP injection into the ileum of C57BL/6J mice. Scale bars, 1 mm. Images representative of n = 3. b, c, Immunofluorescence images of the caudal brainstem 4 days post PRV injection into the: b, ileum (PRV–RFP) and duodenum (PRV–GFP); or c, ileum (PRV–RFP) and colon (PRV–GFP) of C57BL/6J mice. Dashed lines outline the borders of the DMV. Image representative of n = 3. d, Immunofluorescence images of brainstem sensory nuclei from VGLUT2L10GFP mice 4 days post injection of PRV–RFP using with anti-GFP and anti-RFP antibodies. e, Immunofluorescence images of brain slices from C57BL/6J mice treated with PBS or streptomycin. Images representative of n = 9. f, Immunofluorescence images of NTS and AP from C57BL/6J GF mice stained with anti-cFos antibody (black). Image representative of n = 3. g, Number of TRAP+ cells in the AP and NTS of FosTRAP2:tdTomato mice treated with streptomycin (n = 5) or PBS (n = 4), given 4-OHT 24 h later. h, Immunofluorescence images of brain slices showing TRAP+ cells in the AP and NTS of FosTRAP2:tdTomato mice treated with PBS (left) or streptomycin (right), given 4-OHT 24 h later. Images representative of n = 4. i, j, Immunofluorescence images of the NTS and AP stained using anti-cFos antibody (black) (i), and number of cFos+ neurons in the AP, NTS and SolG (j), 24 h post streptomycin and concomitant treatment with PBS (n = 5) or tributyrin (n = 4) in C57BL/6J mice. Images representative of PBS (n = 4), tributyrin (n = 5). b–f, h, i, Scale bars, 50 μm. g, j, Means ± s.d. g, j, Two-tailed unpaired t-test.
Extended Data Fig. 9 Targeting of peripheral neurons using different Cre mouse lines.
a, b, Immunofluorescence images of a, NG, CG-SMG and DRG or b, myenteric plexus and villi in the ileum from Phox2btdTomato:RiboTag, SNStdTomato:RiboTag and Nav1.8tdTomato:RiboTag mice. Images representative of n = 3. In b, red dashed lines highlight the villi structure. c, Number of cFos+ neurons in the CG-SMG of Trpv1Cre− and Trpv1DTA mice treated with PBS (n = 2, n = 3 respectively) or streptomycin (n = 3, n = 3 respectively). d, Immunofluorescence images of CG-SMG from C57BL/6J mice injected with AAV2–hSyn–hM3Dq–mCherry or AAV2–hSyn–hM3Dq–mCherry virus, post administration of 1 mg kg−1 C21. Arrows indicate cFos+ mCherry+ cells. Asterisks indicate other cFos+ cells (possibly haematopoietic cells). e, Percentage of cFos+ among mCherry+ neurons in the CG-SMG of C57BL/6J mice injected with AAV2–hSyn–hM3Dq–mCherry (n = 4) or AAV2–hSyn–hM4Di–mCherry (n = 5 or n = 4) virus, post administration of 1 mg kg−1 or 10 mg kg−1 C21. f, Number of cFos+ neurons in the CG-SMG of C57BL6/J mice injected with AAV2–hSyn–hM3Dq–mCherry (n = 4) or AAV2–hSyn–hM4Di–mCherry virus post administration of 1 mg kg−1 (n = 5) or 10 mg kg−1 (n = 4) C21. g, Number of mCherry+ neurons in the CG-SMG of C57BL/6J mice injected with AAV2–hSyn–hM3Dq–mCherry (n = 4) or AAV2–hSyn–hM4divmCherry post administration of 1 mg kg−1 (n = 5) or 10 mg kg−1 (n = 4) C21. h, Immunofluorescence images of CG-SMG from Nav1.8tdTomato, AviltdTomato and SNStdTomato mice injected with CTB488 in the proximal colon. Arrows indicate neurons that are CTB+ and tdTomato–. i, Number of cFos+ neurons in the CG-SMG of Nav1.8hM4Di mice 6 h post administration with 10 mg kg−1 C21 (n = 3) or saline (n = 3). j, RNAscope in situ hybridization images of L-NG and R-NG for Scn5a (Nav1.5) (red). Images representative of n = 4. k, RNAscope in situ hybridization images of the R-NG for Scn5a (Nav1.5) (red) and Cre from mice injected with AAVrg–pmSyn–Cre–EGFP into the proximal colon. Images representative of n = 2. l, RNAscope in situ hybridization images of the R-NG for Scn5a (Nav1.5) (red) and immunofluorescence for CTB488 from mice with CTB488-injected into the proximal colon. Images representative of n = 2. a, b, d, h, j–l, Scale bars, 50 μm. c, e–g, i, Means ± s.d. i, Two-tailed unpaired t-test. c, e–g, One-way ANOVA with Tukey’s multiple comparisons.
Extended Data Fig. 10 Gut-connected brainstem regions modulate intestinal motility.
a, b, Immunofluorescence images of brain slices highlighting the NTS/AP from a, SNShM4Di and b, Phox2bhM4Di mice treated with C21 at 10 mg kg−1, stained with anti-cFos antibody (black). Images representative of n = 3. c, d, Immunofluorescence images of brain slices highlighting the NTS/AP from c, sham-operated C57BL/6J mice treated with streptomycin, and d, sdVx C57BL/6J mice treated with PBS, stained with anti-cFos antibody (black). Images representative of n = 5. e, Immunofluorescence image of the L-NG post injection of PRV–GFP into LPGi/RVLM of brainstem. Outline highlights NG. Image representative of n = 3. f, g, Immunofluorescence images of f, CG-SMG and g, L-NG after PRV–GFP and CTB594 injection into the proximal colon of sdVx C57BL/6J mice. Image representative of n = 3. Outline highlights NG. h, i, Left, schemes showing injection of AAVrg–FLEX–tdTomato into the NTS of h, VGLUT2Cre and i, SNSChR2 mice. Right, immunofluorescence images of the NG and colonic myenteric plexus. The white dotted outlines in the left-hand images highlight the NG. Images representative of n = 2. i, The magnified image (second from right) shows tdTomato+ fibre surrounding an intrinsic enteric neuron. The brainstem slice (right) shows the absence of Cre activity in the dorsal motor nucleus of the vagus. Images representative of n = 3. j, Left, scheme showing injection into the R-NG. Right, expression of AAV9–Syn–ChrimsonR–tdTomato in L-NG and R-NG. Image representative of n = 3. k, Immunofluorescence cleared-tissue images of the duodenum (left) and caecum (right) of mice injected with AAV9–Syn–ChrimsonR–tdTomato in the R-NG. Scale bars, 500 mm. Image representative of n = 3. l, Scheme showing microbiota-sensitive circuits controlling sympathetic activity to the gut, and potentially to other visceral organs. a–j, Scale bars, 50 μm.
Supplementary information
Supplementary Figure 1
Annotated snapshots of Supplementary Videos 1-11.
Supplementary Information
Supplementary Information 1: Role of Microbiota-Modulated Epithelial Factors in CG-SMG Activation. Manipulation of the epithelial microbiota-modulated signalling molecules revealed that the neuropeptides GLP-1 and PYY can modulate the activation state of gut sympathetic neurons and, in the case of PYY, may contribute to microbial regulation of gut sympathetic activity.
Supplementary Information
Supplementary Information 2: CG-SMG Intrinsic or Viscerofugal Modulation of CG-SMG Activation. Anterograde/retrograde tracing, cleared tissue imaging, chemical sympathectomy, and analysis of iEAN activation markers were utilized to determine that CG-SMG intrinsic or viscerofugal modulation unlikely lead to CG-SMG activation in the context of microbial depletion.
Supplementary Information
Supplementary Information 3: Ruling out hM4Di mediated activation of CG-SMG neurons. To rule out a role for hM4Di-mediated activation of CG-SMG neurons, specific CG-SMG chemogenetic activation or inhibition was performed and an expected robust increase in cFos expression was observed only in hM3Dq+ and not hM4Di+ sympathetic neurons after C21 administration.
Video 1
AdipoClear whole mount of colon stained with anti-RFP showing tdTomato+ fibres from the CG-SMG 2 weeks post AAVrg-CAG-FLEX-tdTomato injection. Representative of n=2.
Video 2
AdipoClear whole mount of cecum stained with anti-RFP showing tdTomato+ fibres from the CG-SMG 2 weeks post AAVrg-CAG-FLEX-tdTomato injection. Representative of n=2.
Video 3
3DISCO whole mount of spinal cord with neurons retrograde traced from the CG-SMG with CTB555. Representative of n=3.
Video 4
calAdipoClear whole mount of spinal cord stained with anti-RFP (red) and anti-TH (green), showing the CG-SMG 4 days post PRV infection in the ileum. Representative of n=3.
Video 5
calAdipoClear whole mount of spinal cord stained with anti-RFP (red) and anti-TH (white) 4 days post PRV infection in the ileum. Representative of n=3.
Video 6
AdipoClear whole mount of brain stained with anti-RFP (red) 3 and 4 days post PRV infection in the ileum, showing the brainstem. Representative of n=3.
Video 7
AdipoClear whole mount of duodenum stained with anti-RFP (white) 2 weeks post injection of AAV9-Syn-ChrimsonR-tdTomato into the RNG. Representative of n=3.
Video 8
AdipoClear whole mount of ileum stained with anti-RFP (white) 2 weeks post injection of AAV9-Syn-ChrimsonR-tdTomato into the RNG. Representative of n=3.
Video 9
AdipoClear whole mount of colon stained with anti-RFP (white) 2 weeks post injection of AAV9-Syn-ChrimsonR-tdTomato into the RNG. Zoomed in on the epithelium. Representative of n=3.
Video 10
AdipoClear whole mount of colon stained with anti-RFP (white) 2 weeks post injection of AAV9-Syn-ChrimsonR-tdTomato into the RNG. Zoomed in on the epithelium. Representative of n=3.
Video 11
AdipoClear whole mount of colon stained with anti-RFP (white) 2 weeks post injection of AAV9-Syn-ChrimsonR-tdTomato into the RNG. Zoomed in on a single semilunar fold. Representative of n=3.
Supplementary Table 1
Description of Cre mouse lines used.
Source data
Rights and permissions
About this article
Cite this article
Muller, P.A., Schneeberger, M., Matheis, F. et al. Microbiota modulate sympathetic neurons via a gut–brain circuit. Nature 583, 441–446 (2020). https://doi.org/10.1038/s41586-020-2474-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2474-7
This article is cited by
-
Microbiota-derived acetate attenuates neuroinflammation in rostral ventrolateral medulla of spontaneously hypertensive rats
Journal of Neuroinflammation (2024)
-
The gut microbiota modulate locomotion via vagus-dependent glucagon-like peptide-1 signaling
npj Biofilms and Microbiomes (2024)
-
Akkermansia muciniphila Improves Depressive-Like Symptoms by Modulating the Level of 5-HT Neurotransmitters in the Gut and Brain of Mice
Molecular Neurobiology (2024)
-
Butterflies in the gut: the interplay between intestinal microbiota and stress
Journal of Biomedical Science (2023)
-
Akkermansia muciniphila, which is enriched in the gut microbiota by metformin, improves cognitive function in aged mice by reducing the proinflammatory cytokine interleukin-6
Microbiome (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.