Abstract
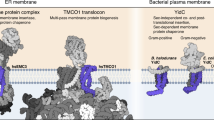
The endoplasmic reticulum (ER) membrane complex (EMC) cooperates with the Sec61 translocon to co-translationally insert a transmembrane helix (TMH) of many multi-pass integral membrane proteins into the ER membrane, and it is also responsible for inserting the TMH of some tail-anchored proteins1,2,3. How EMC accomplishes this feat has been unclear. Here we report the first, to our knowledge, cryo-electron microscopy structure of the eukaryotic EMC. We found that the Saccharomyces cerevisiae EMC contains eight subunits (Emc1–6, Emc7 and Emc10), has a large lumenal region and a smaller cytosolic region, and has a transmembrane region formed by Emc4, Emc5 and Emc6 plus the transmembrane domains of Emc1 and Emc3. We identified a five-TMH fold centred around Emc3 that resembles the prokaryotic YidC insertase and that delineates a largely hydrophilic client protein pocket. The transmembrane domain of Emc4 tilts away from the main transmembrane region of EMC and is partially mobile. Mutational studies demonstrated that the flexibility of Emc4 and the hydrophilicity of the client pocket are required for EMC function. The EMC structure reveals notable evolutionary conservation with the prokaryotic insertases4,5, suggests that eukaryotic TMH insertion involves a similar mechanism, and provides a framework for detailed understanding of membrane insertion for numerous eukaryotic integral membrane proteins and tail-anchored proteins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM 3D map of the S. cerevisiae EMC complex has been deposited at the Electron Microscopy Data Bank (EMDB) database with accession code EMD-21587. The corresponding atomic model was deposited at the RCSB Protein Data Bank (PDB) with accession code 6WB9. The TMT mass spectrometry data and the real-space correlation coefficients of all residues with experimental densities data are provided in Supplementary Tables 1–3. The uncropped SDS–PAGE gels used in Figs. 1a, 3g and Extended Data Fig. 6b can be found in Supplementary Fig. 1.
References
Shurtleff, M. J. et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife 7, e37018 (2018).
Chitwood, P. J., Juszkiewicz, S., Guna, A., Shao, S. & Hegde, R. S. EMC is required to initiate accurate membrane protein topogenesis. Cell 175, 1507–1519 (2018).
Guna, A., Volkmar, N., Christianson, J. C. & Hegde, R. S. The ER membrane protein complex is a transmembrane domain insertase. Science 359, 470–473 (2018).
Chen, Y. & Dalbey, R. E. Oxa1 superfamily: new members found in the ER. Trends Biochem. Sci. 43, 151–153 (2018).
Anghel, S. A., McGilvray, P. T., Hegde, R. S. & Keenan, R. J. Identification of oxa1 homologs operating in the eukaryotic endoplasmic reticulum. Cell Rep. 21, 3708–3716 (2017).
Fry, M. Y. & Clemons, W. M. Jr. Complexity in targeting membrane proteins. Science 359, 390–391 (2018).
Richard, M., Boulin, T., Robert, V. J., Richmond, J. E. & Bessereau, J. L. Biosynthesis of ionotropic acetylcholine receptors requires the evolutionarily conserved ER membrane complex. Proc. Natl Acad. Sci. USA 110, E1055–E1063 (2013).
Coelho, J. P. L. et al. A network of chaperones prevents and detects failures in membrane protein lipid bilayer integration. Nat. Commun. 10, 672 (2019).
Tian, S. et al. Proteomic analysis identifies membrane proteins dependent on the ER membrane protein complex. Cell Rep. 28, 2517–2526 (2019).
Hiramatsu, N., Tago, T., Satoh, T. & Satoh, A. K. ER membrane protein complex is required for the insertions of late-synthesized transmembrane helices of Rh1 in Drosophila photoreceptors. Mol. Biol. Cell 30, 2859–2942 (2019).
Bagchi, P., Inoue, T. & Tsai, B. EMC1-dependent stabilization drives membrane penetration of a partially destabilized non-enveloped virus. eLife 5, e21470 (2016).
Christianson, J. C. et al. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 14, 93–105 (2011).
Jonikas, M. C. et al. Comprehensive characterization of genes required for protein folding in the endoplasmic reticulum. Science 323, 1693–1697 (2009).
Lahiri, S. et al. A conserved endoplasmic reticulum membrane protein complex (EMC) facilitates phospholipid transfer from the ER to mitochondria. PLoS Biol. 12, e1001969 (2014).
Wideman, J. G. The ubiquitous and ancient ER membrane protein complex (EMC): tether or not? F1000Res. 4, 624 (2015).
Luo, W.-J., Gong, X.-H. & Chang, A. An ER membrane protein, Sop4, facilitates ER export of the yeast plasma membrane [H+]ATPase, Pma1. Traffic 3, 730–739 (2002).
Bircham, P. W. et al. Secretory pathway genes assessed by high-throughput microscopy and synthetic genetic array analysis. Mol. Biosyst. 7, 2589–2598 (2011).
Kudze, T., Mendez-Dorantes, C., Jalloh, C. S. & McClellan, A. J. Evidence for interaction between Hsp90 and the ER membrane complex. Cell Stress Chaperones 23, 1101–1115 (2018).
Pausch, P. et al. Co-translational capturing of nascent ribosomal proteins by their dedicated chaperones. Nat. Commun. 6, 7494 (2015).
Baßler, J. et al. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J. Cell Biol. 210, 169–170 (2015).
Tang, X. et al. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc. Natl Acad. Sci. USA 109, 3287–3292 (2012).
Kedrov, A. et al. Structural dynamics of the YidC:ribosome complex during membrane protein biogenesis. Cell Rep. 17, 2943–2954 (2016).
Wickles, S. et al. A structural model of the active ribosome-bound membrane protein insertase YidC. eLife 3, e03035 (2014).
Kohler, R. et al. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol. Cell 34, 344–353 (2009).
Kumazaki, K. et al. Structural basis of Sec-independent membrane protein insertion by YidC. Nature 509, 516–520 (2014).
Xin, Y. et al. Structure of YidC from Thermotoga maritima and its implications for YidC-mediated membrane protein insertion. FASEB J. 32, 2411–2421 (2018).
Kumazaki, K. et al. Crystal structure of Escherichia coli YidC, a membrane protein chaperone and insertase. Sci. Rep. 4, 7299 (2014).
Xie, K., Kiefer, D., Nagler, G., Dalbey, R. E. & Kuhn, A. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry 45, 13401–13408 (2006).
Sachelaru, I. et al. YidC occupies the lateral gate of the SecYEG translocon and is sequentially displaced by a nascent membrane protein. J. Biol. Chem. 288, 16295–16307 (2013).
Lin, D. L. et al. The ER membrane protein complex promotes biogenesis of dengue and Zika virus non-structural multi-pass transmembrane proteins to support infection. Cell Rep. 27, 1666–1674 (2019).
Funakoshi, M. & Hochstrasser, M. Small epitope-linker modules for PCR-based C-terminal tagging in Saccharomyces cerevisiae. Yeast 26, 185–192 (2009).
Kammers, K., Cole, R. N., Tiengwe, C. & Ruczinski, I. Detecting significant changes in protein abundance. EuPA Open Proteom. 7, 11–19 (2015).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Amunts, A. et al. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485–1489 (2014).
Acknowledgements
Cryo-EM images were collected in the David Van Andel Advanced Cryo-Electron Microscopy Suite at Van Andel Research Institute. We thank G. Zhao and X. Meng for assistance with data collection and D. Nadziejka for critical reading of the manuscript. This work was supported by the US National Institutes of Health (NIH) (R01 CA231466 to H.L.) and Van Andel Institute (to H.L.).
Author information
Authors and Affiliations
Contributions
L.B. and H.L. conceived and designed experiments. L.B. performed most of the experiments. Q.Y., X.F. and A.K. helped with sample preparation and functional assays. L.B. and H.L. analysed the data and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Friedrich Förster and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Data processing and validation of cryo-EM micrographs and 3D reconstruction.
a, Gel filtration profile of the EMC complex. This experiment was repeated more than five times with similar results. b, c, Representative electron micrograph and selected reference-free 2D class averages of the EMC. A total of 4,260 micrographs were recorded with similar quality. d, Cryo-EM data-processing procedure. e, Gold-standard Fourier shell correlations of two independent half maps with or without mask, and with randomized phases, and the validation correlation curves of the atomic model by comparing the model with the final map or with the two half maps. f, Local-resolution map of the 3D map. g, Angular distribution of particles used in the final reconstruction of the 3D map.
Extended Data Fig. 2 Protein abundance and localization of nine putative EMC clients in wild-type and Emc3-knockout yeast strains.
The eGFP is appended to the C termini of the genes. Scale bar, 10 μm. This experiment was repeated three times with similar results.
Extended Data Fig. 3 Cryo-EM 3D density map of the EMC.
a–d, The surface-rendered map is shown in front view (a), left side view (b), right side view (c), back view (d), bottom (lumenal) view (e), and top (cytosolic) view (f). Maps are coloured by individual subunits.
Extended Data Fig. 4 The fitting of the atomic model and the 3D map in selected regions.
3D density map and atomic model of selected regions in each of the eight EMC subunits, as well as the densities of atomic models of the two phospholipid molecules. C-ter, C-terminal domain; HH, horizontal helix; N-ter, N-terminal domain.
Extended Data Fig. 5 Structure of the lumenal and cytosolic regions of the yeast EMC.
a, Structure of the EMC lumenal region shown in front side and bottom (lumenal) views. The interface area between the C-terminal loop of Emc4 and the NTD2 of Emc1 is outlined by a red rectangle. The dotted black area marks the NTD2 of Emc1, which is an eight-bladed β-propeller. b, Superposition of the NTD2 β-propeller of Emc1 with the structure of a fungus chaperone protein Sqt1 (PDB code 4ZN4). c, Enlarged view of the red-outlined region in a. d, Structure of the EMC cytosolic region in top (cytosolic) and front side views. Emc2 as the organizing centre is shown in cartoon, and the cytosolic domains of Emc3, Emc4 and Emc5 are shown as cylinders.
Extended Data Fig. 6 In vitro binding assays between the purified EMC and the TOM complex.
a, Gel filtration profiles of the EMC alone, the TOM complex alone, and the mixture of the EMC–TOM complexes. No peak corresponding to the assembly of the EMC–TOM complex was observed. The experiment was repeated three times yielding similar results. b, Peak fractions of the EMC–TOM mixture in a were checked by the Coomassie blue-stained SDS–PAGE gel. The band densities suggest that the peak is simply an overlap of the unbound and separate EMC and TOM. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 7 The 3D EM map of the EMC surface rendered at a low display threshold.
The bound lipids/detergents surrounding the transmembrane region of the EMC complex are visible in this low-threshold display. The atomic model in cartoon is superimposed on the 3D map. Note that the horizontal helix of Emc1 is at the ER lumen–membrane boundary.
Extended Data Fig. 8 Structural comparison between yeast EMC and E. coli YidC.
a, Structure of EMC in cartoon. b, Structure of E. coli YidC in cartoon (PDB code 3WVF). c, Superposed structures of EMC (colour) and YidC (dark grey).
Extended Data Fig. 9 Comparisons of protein abundance, localization, and growth of the mutant yeast strains with the wild-type cells.
a, Protein abundance and localization of two putative EMC clients (Mrh1 and Fet3) in wild-type and EMC3(K26L) mutant yeast strains. The eGFP is appended to the C termini of the genes. b, Growth experiments of yeast strains containing Emc4 linker loop truncations. The three truncations were Emc4(∆56–60), Emc4(∆51–60), and Emc4(∆46–60). Experiments in a and b were repeated three times with similar results.
Supplementary information
Supplementary Figure 1
Uncropped SDS-PAGE gels used for preparing Fig. 1a, Fig. 3g, and Extended Data Fig. 6b.
Supplementary Table 1 | EMC dependent proteins identified by TMT mass spectrometry
The N positions were either from literature as cited or as predicted by TMHMM Server V. 2.0.
Supplementary Table 3 | The list of real-space correlation coefficients of all residues with experimental densities
The correlations were calculated for each amino acid between the atomic model and the 3D density map by PHENIX-MolProbity.
Supplementary Video 1 | Cryo-EM structure of the EMC complex
A surface rendering of the 3D map, superimposed with the atomic model, is shown rotating around a vertical axis by 360°, followed by another 360° rotation around a horizontal axis. The eight subunits are individually colored. The red densities are from the five resolved N-glycans.
Rights and permissions
About this article
Cite this article
Bai, L., You, Q., Feng, X. et al. Structure of the ER membrane complex, a transmembrane-domain insertase. Nature 584, 475–478 (2020). https://doi.org/10.1038/s41586-020-2389-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2389-3
This article is cited by
-
EMC rectifies the topology of multipass membrane proteins
Nature Structural & Molecular Biology (2024)
-
EMC chaperone–CaV structure reveals an ion channel assembly intermediate
Nature (2023)
-
Molecular pathway of mitochondrial preprotein import through the TOM–TIM23 supercomplex
Nature Structural & Molecular Biology (2023)
-
Expulsion mechanism of the substrate-translocating subunit in ECF transporters
Nature Communications (2023)
-
The mechanisms of integral membrane protein biogenesis
Nature Reviews Molecular Cell Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.