Abstract
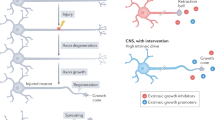
Grafts of spinal-cord-derived neural progenitor cells (NPCs) enable the robust regeneration of corticospinal axons and restore forelimb function after spinal cord injury1; however, the molecular mechanisms that underlie this regeneration are unknown. Here we perform translational profiling specifically of corticospinal tract (CST) motor neurons in mice, to identify their ‘regenerative transcriptome’ after spinal cord injury and NPC grafting. Notably, both injury alone and injury combined with NPC grafts elicit virtually identical early transcriptomic responses in host CST neurons. However, in mice with injury alone this regenerative transcriptome is downregulated after two weeks, whereas in NPC-grafted mice this transcriptome is sustained. The regenerative transcriptome represents a reversion to an embryonic transcriptional state of the CST neuron. The huntingtin gene (Htt) is a central hub in the regeneration transcriptome; deletion of Htt significantly attenuates regeneration, which shows that Htt has a key role in neural plasticity after injury.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
RNA-seq data have been deposited in the Gene Expression Omnibus (GEO) repository (www.ncbi.nlm.nih.gov/geo), accession number GSE126957.
References
Kadoya, K. et al. Spinal cord reconstitution with homologous neural grafts enables robust corticospinal regeneration. Nat. Med. 22, 479–487 (2016).
Nathan, P. W. Effects on movement of surgical incisions into the human spinal cord. Brain 117, 337–346 (1994).
Friedli, L. et al. Pronounced species divergence in corticospinal tract reorganization and functional recovery after lateralized spinal cord injury favors primates. Sci. Transl. Med. 7, 302ra134 (2015).
Oudega, M. & Perez, M. A. Corticospinal reorganization after spinal cord injury. J. Physiol. (Lond.) 590, 3647–3663 (2012).
Liu, K. et al. PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat. Neurosci. 13, 1075–1081 (2010).
Jin, D. et al. Restoration of skilled locomotion by sprouting corticospinal axons induced by co-deletion of PTEN and SOCS3. Nat. Commun. 6, 8074 (2015).
Coumans, J. V. et al. Axonal regeneration and functional recovery after complete spinal cord transection in rats by delayed treatment with transplants and neurotrophins. J. Neurosci. 21, 9334–9344 (2001).
Doyle, J. P. et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135, 749–762 (2008).
Lu, P. et al. Long-distance growth and connectivity of neural stem cells after severe spinal cord injury. Cell 150, 1264–1273 (2012).
Kumamaru, H. et al. Generation and post-injury integration of human spinal cord neural stem cells. Nat. Methods 15, 723–731 (2018).
Heiman, M., Kulicke, R., Fenster, R. J., Greengard, P. & Heintz, N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP). Nat. Protocols 9, 1282–1291 (2014).
Groh, A. et al. Cell-type specific properties of pyramidal neurons in neocortex underlying a layout that is modifiable depending on the cortical area. Cereb. Cortex 20, 826–836 (2010).
Lein, E. S. et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007).
Lodato, S. et al. Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat. Neurosci. 17, 1046–1054 (2014).
Fink, K. L., Strittmatter, S. M. & Cafferty, W. B. Comprehensive corticospinal labeling with μ-crystallin transgene reveals axon regeneration after spinal cord trauma in ngr1 -/- mice. J. Neurosci. 35, 15403–15418 (2015).
Woodworth, M. B., Greig, L. C., Kriegstein, A. R. & Macklis, J. D. SnapShot: cortical development. Cell 151, 918–918.e1 (2012).
Karow, M. et al. Reprogramming of pericyte-derived cells of the adult human brain into induced neuronal cells. Cell Stem Cell 11, 471–476 (2012).
Arlotta, P. et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 (2005).
Hollis, E. R. II, Jamshidi, P., Löw, K., Blesch, A. & Tuszynski, M. H. Induction of corticospinal regeneration by lentiviral trkB-induced Erk activation. Proc. Natl Acad. Sci. USA 106, 7215–7220 (2009).
Park, K. K. et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science 322, 963–966 (2008).
Liu, Y. et al. A sensitized IGF1 treatment restores corticospinal axon-dependent functions. Neuron 95, 817–833 (2017).
Qiu, J. et al. Spinal axon regeneration induced by elevation of cyclic AMP. Neuron 34, 895–903 (2002).
He, Z. & Jin, Y. Intrinsic control of axon regeneration. Neuron 90, 437–451 (2016).
Sekine, Y., Siegel, C. S., Sekine-Konno, T., Cafferty, W. B. J. & Strittmatter, S. M. The nociceptin receptor inhibits axonal regeneration and recovery from spinal cord injury. Sci. Signal. 11, eaao4180 (2018).
Vodrazka, P. et al. The semaphorin 4D–plexin-B signalling complex regulates dendritic and axonal complexity in developing neurons via diverse pathways. Eur. J. Neurosci. 30, 1193–1208 (2009).
Hayashi, N., Miyata, S., Yamada, M., Kamei, K. & Oohira, A. Neuronal expression of the chondroitin sulfate proteoglycans receptor-type protein-tyrosine phosphatase β and phosphacan. Neuroscience 131, 331–348 (2005).
Maden, C. H. et al. NRP1 and NRP2 cooperate to regulate gangliogenesis, axon guidance and target innervation in the sympathetic nervous system. Dev. Biol. 369, 277–285 (2012).
Samuels, I. S., Saitta, S. C. & Landreth, G. E. MAP'ing CNS development and cognition: an ERKsome process. Neuron 61, 160–167 (2009).
Poplawski, G. et al. Adult rat myelin enhances axonal outgrowth from neural stem cells. Sci. Transl. Med. 10, eaal2563 (2018).
Dulin, J. N. et al. Injured adult motor and sensory axons regenerate into appropriate organotypic domains of neural progenitor grafts. Nat. Commun. 9, 84 (2018).
Adler, A. F., Lee-Kubli, C., Kumamaru, H., Kadoya, K. & Tuszynski, M. H. Comprehensive monosynaptic rabies virus mapping of host connectivity with neural progenitor grafts after spinal cord injury. Stem Cell Reports 8, 1525–1533 (2017).
Koffler, J. et al. Biomimetic 3D-printed scaffolds for spinal cord injury repair. Nat. Med. 25, 263–269 (2019).
Belin, S. et al. Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron 86, 1000–1014 (2015).
McKinstry, S. U. et al. Huntingtin is required for normal excitatory synapse development in cortical and striatal circuits. J. Neurosci. 34, 9455–9472 (2014).
Phillips, O. et al. The corticospinal tract in Huntington’s disease. Cereb. Cortex 25, 2670–2682 (2015).
Zeitlin, S., Liu, J. P., Chapman, D. L., Papaioannou, V. E. & Efstratiadis, A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nat. Genet. 11, 155–163 (1995).
Dietrich, P., Johnson, I. M., Alli, S. & Dragatsis, I. Elimination of huntingtin in the adult mouse leads to progressive behavioral deficits, bilateral thalamic calcification, and altered brain iron homeostasis. PLoS Genet. 13, e1006846 (2017).
Wang, G., Liu, X., Gaertig, M. A., Li, S. & Li, X. J. Ablation of huntingtin in adult neurons is nondeleterious but its depletion in young mice causes acute pancreatitis. Proc. Natl Acad. Sci. USA 113, 3359–3364 (2016).
Landles, C. & Bates, G. P. Huntingtin and the molecular pathogenesis of Huntington’s disease. EMBO Rep. 5, 958–963 (2004).
Butler, R. & Bates, G. P. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat. Rev. Neurosci. 7, 784–796 (2006).
Duyao, M. P. et al. Inactivation of the mouse Huntington’s disease gene homolog Hdh. Science 269, 407–410 (1995).
Ross, C. A. et al. Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat. Rev. Neurol. 10, 204–216 (2014).
Marcora, E. & Kennedy, M. B. The Huntington’s disease mutation impairs Huntingtin’s role in the transport of NF-κB from the synapse to the nucleus. Hum. Mol. Genet. 19, 4373–4384 (2010).
Trushina, E. et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol. Cell. Biol. 24, 8195–8209 (2004).
Dragatsis, I., Levine, M. S. & Zeitlin, S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 26, 300–306 (2000).
Fouad, K., Pedersen, V., Schwab, M. E. & Brösamle, C. Cervical sprouting of corticospinal fibers after thoracic spinal cord injury accompanies shifts in evoked motor responses. Curr. Biol. 11, 1766–1770 (2001).
Weidner, N., Ner, A., Salimi, N. & Tuszynski, M. H. Spontaneous corticospinal axonal plasticity and functional recovery after adult central nervous system injury. Proc. Natl Acad. Sci. USA 98, 3513–3518 (2001).
Bareyre, F. M. et al. The injured spinal cord spontaneously forms a new intraspinal circuit in adult rats. Nat. Neurosci. 7, 269–277 (2004).
Patel, A. et al. AxonTracer: a novel ImageJ plugin for automated quantification of axon regeneration in spinal cord tissue. BMC Neurosci. 19, 8 (2018).
Heiman, M. et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748 (2008).
Auer, P. L. & Doerge, R. W. Statistical design and analysis of RNA sequencing data. Genetics 185, 405–416 (2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Plaisier, S. B., Taschereau, R., Wong, J. A. & Graeber, T. G. Rank–rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nucleic Acids Res. 38, e169 (2010).
Acknowledgements
This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Veterans Administration (The Gordon Mansfield Consortium for Spinal Cord Regeneration), the NIH (NS09881 and EB014986) and the Gerbic Family Foundation. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691 to G.C.). We appreciate the assistance of J. Dulin.
Author information
Authors and Affiliations
Contributions
G.H.D.P. and M.H.T. conceptualized the study; G.H.D.P., R.K., G.C. and M.H.T. performed analysis of transcriptomic data; G.H.D.P., P.C. and R.L. performed data validation; G.H.D.P., R.L., N.M. and P.C. performed formal analysis; G.H.D.P., R.L., N.M., P.C., E.V.N. and J.M.M. performed investigation; G.H.D.P., N.M., P.C. and R.L. performed experiments and analysis for Fig.1; G.H.D.P., R.K. and G.C. performed experiments and data analysis and validation for Fig. 2, Extended Data Figs. 2, 6, 7; E.V.N. and J.M.M. performed experiments and analysis for Fig. 3; P.L. performed experiments for Extended Data Fig. 1; G.H.D.P. performed analysis for Extended Data Fig. 3; G.H.D.P. performed experiments and analysis for Extended Data Fig. 5; G.H.D.P. performed experiment and analysis for Extended Data Fig. 7; I.D., B.Z., G.C. and M.H.T. provided materials used for experiments; G.H.D.P., R.K. and G.C. performed data curation; G.H.D.P. and M.H.T. wrote the original draft; G.H.D.P., M.H.T., R.K. and G.C. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 TRAP of regenerating corticospinal neurons.
a, b, Coronal cortical section of transgenic BAC–TRAP mice expressing eGFP–L10A under the Glt25d2 promoter primarily in neurons (NeuN) of cortical layer 5b. b, Confocal z-stack projection of 15-μm thickness showing colocalization of eGFP–L10A and NeuN within pyramidal neurons. c, d, Retrogradely labelled CST neurons (CTB) projecting to the cervical spinal cord (C4–C8) grey matter colabel with eGFP–L10A, indicating that about 50% of TRAPed mRNA molecules are derived from corticospinal neurons. d, Confocal z-stack projection of 10-μm thickness showing colocalization of eGFP–L10A and CTB within pyramidal neurons. n = 3 biological replicates, which showed comparable results. e, Experimental timeline for purification of TRAPed mRNA molecules exclusively from cortical-layer-5 neurons.
Extended Data Fig. 2 Sample Pearson correlation identifies unique populations of mRNAs derived from CST neurons (TRAPed samples) and from whole motor cortex (unbound samples) after TRAP.
mRNA species from all conditions are more similar to each other (TRAPed samples) than total RNA collected from the motor cortex (unbound samples), indicating that TRAP enriched for a uniquely defined mRNA population that is distinct from whole cortex samples. Samples are arranged by hierarchical clustering.
Extended Data Fig. 3 In situ hybridization data from the Allen Mouse Brain Atlas confirms layer-5b-specific mRNA expression of genes highly enriched by TRAP from intact Glt25d2-eGFP-L10a mice.
a, Fold change of most highly enriched mRNA transcripts in the corticospinal cell population (TRAPed samples) compared to mRNA from whole motor cortex (unbound samples) in the intact state. n = 3 biological replicates per condition. b, In situ hybridization (ISH) data (Allen Mouse Brain Atlas; https://mouse.brain-map.org/) of most highly enriched genes (from a) confirms cell-type-specific enrichment of TRAPed mRNA expression in layer-5b neurons of the adult mouse motor cortex.
Extended Data Fig. 4 Neuronal genes are enriched in the TRAPed samples from layer-5b neurons.
a, Neuronal marker genes are enriched in the TRAPed samples derived from layer-5b neurons. b, Astrocyte- and oligodendrocyte-specific markers are expressed at a higher level in the whole motor cortex (unbound samples). n = 3 biological replicates per condition. Data are mean ± s.d.
Extended Data Fig. 5 Verification of RNA-seq data via immunolabelling for ATG7, SOX9 and PAX2.
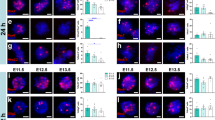
Immunohistological staining of selected proteins in layer 5b of the motor cortex of Glt25d2-eGFP-L10a transgenic mice. a, b, eGFP–L10A staining is shown in green and SOX9 staining is shown in red in intact mice (a) or in regenerating CST 10 days after injury with E12 NPC graft (b). c, Sox9 mRNA expression after TRAP and RNA-seq. Data are normalized to those from intact mice. ****P = 3.2 × 10−7, two-tailed t-test. d, e, eGFP–L10A staining is shown in green and PAX2 staining is shown in red in intact mice (d) or in regenerating CST 10 days after injury with E12 NPC graft (e). f, Pax2 mRNA expression after TRAP and RNA-seq. Data are normalized to those from intact mice. **P = 0.0057, two-tailed t-test. g, h, eGFP–L10A staining is shown in green and ATG7 staining is shown in red in intact mice (g) or in regenerating CST 10 days after injury with E12 NPC graft (h). i, Atg7 mRNA expression after TRAP and RNA-seq. Data are normalized to those from intact mice. ****P = 3.0 × 10−6, two-tailed t-test. n = 3 biological replicates per condition. Data are mean ± s.d.
Extended Data Fig. 6 Heat map of the top 1,000 significantly differentially regulated transcripts (FDR < 0.1) upon injury and upon regeneration, relative to intact mice, with individual gene names.
Heat map of the top 1,000 significantly differentially regulated transcripts (FDR < 0.1, Benjamini–Hochberg, two-sided) upon injury and upon regeneration, relative to the intact state. Red, increased expression; green, reduced expression. The intensity of the colour reflects the degree of gene regulation compared to the intact state. Transcripts are arranged by hierarchical clustering. n = 2 biological replicates for day-10 non-regenerating, and n = 3 biological replicates for all other conditions. Genes are listed from top to bottom. Black and grey boxes next to heat map indicate gene lists (columns 1 to 7 from left to right).
Extended Data Fig. 7 Heat map of genes enriched in the Gene Ontology analysis (Ingenuity Pathway Analysis software).
Heat map of Gene Ontology (GO)-term-associated significantly differentially regulated transcripts (FDR < 0.1, Benjamini–Hochberg, two-sided) upon injury and upon regeneration, relative to the intact state, at 10, 14 and 21 days after injury. Red, increased expression; green, reduced expression. The intensity of the colour reflects the degree of gene regulation compared to the intact state. Transcripts are arranged by GO-term clustering (Ingenuity Pathway Analysis software). GO clusters listed on the right are indicated via colour code on the left side of the heat map. Many common genes have increased in expression in both non-regenerating and regenerating groups 10 days after injury. However, by day 14, these injury-induced changes are diminishing in the absence of a graft but are sustained in the presence of a graft, into which regeneration occurs. By day 21 after injury, many genes have returned to baseline levels in non-regenerating CST neurons, whereas they remain highly upregulated in regenerating CST neurons. The GO categories are derived from Figs. 2, 3. n = 2 biological replicates for day-10 non-regenerating, and n = 3 biological replicates for all other conditions.
Extended Data Fig. 8 The regeneration gene-expression profile is sustained at 21 days only with an NPC graft, and not with a non-stimulatory MSC graft, and correlates with neuronal developmental transcriptomes.
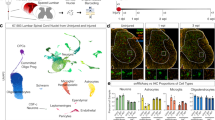
a, Number of significantly differentially expressed genes of non-regenerating (no graft and MSC graft) and regenerating (NPC graft) CST neurons relative to intact CST neurons (FDR < 0.1, Benjamini–Hochberg, two-sided) at 21 days after injury. Red, increased expression; green, reduced expression. b, Heat map of the top 1,000 significantly differentially regulated transcripts (FDR < 0.1) upon injury and upon regeneration, relative to the intact state, at 21 days after injury. Red, increased expression; green, reduced expression. The intensity of the colour reflects the degree of gene regulation compared to the intact state. Transcripts are arranged by hierarchical clustering. The non-regenerating conditions (no graft and MSC graft) show gene-expression patterns similar to one another; and, in both cases, gene expression is not sustained in comparison to the regenerating condition (NPC graft). n = 3 biological replicates. c, Differentially expressed genes (FDR < 0.1) of a previous neuronal development study (GSE2039) for different time points against P14 (log-transformed fold change) was compared to CST data against the intact condition. Numbers indicate the Pearson correlation coefficient of the two datasets. Colour indicates the magnitude of the correlation coefficient (side bar). Values in parentheses are statistical significance (P values) of the correlation coefficients. n = 2 biological replicates for day-10 non-regenerating and n = 3 biological replicates for all other conditions. d–f, Overlapping signalling pathways of regenerating versus non-regenerating CST neurons (FDR < 0.1) using Ingenuity Pathway Analysis software at 10 (d), 14 (e) and 21 (f) days after injury. The network of overlapping signalling pathways shows each pathway as a single ‘node’ that is coloured proportionally to the right-tailed Fisher’s exact test P value; darker red, more significant. Individual scaling of P values is listed at the bottom of each graph. A line connects any two pathways when there is at least one dataset molecule in common between them. Pathways of similar function are clustered together (dashed blue outlines). n = 2 biological replicates for day-10 non-regenerating and n = 3 biological replicates for all other conditions.
Extended Data Fig. 9 AAV8-tdTomato-Cre expression completely abolishes detection of HTT protein six weeks after injection.
AAV8-dependent Cre delivery to Httflox/flox mice was used to conditionally knock out HTT expression in corticospinal neurons six weeks before injury. a–c, Low magnification of right motor cortex six weeks after AAV8-tdTomato-Cre injection indicates successful tdTomato (red) expression in motor cortex and reduction in HTT protein (black or blue) in the infected area. d, e, High-magnification images showing cortical neurons with baseline HTT protein expression outside of infected areas (d) and after HTT deletion in cortical neurons that are transfected with AAV8-tdTomato-Cre (e). f, AAV8-tdTomato-Cre-infected cortical neurons co-express tdTomato (red) and Cre (green). n = 3 biological replicates, with comparable outcomes.
Extended Data Fig. 10 Lesioned corticospinal axons do not regenerate in the absence of an NPC graft.
a, CST axons labelled with the anterograde axonal tracer BDA do not regenerate in the absence of a growth-stimulating NPC graft. The GFAP label indicates glial activation around the lesion site. b, However, CST axons in mice without grafts do sprout into spared host grey matter underlying the lesion, consistent with previous reports46,47,48. In the box plot, centre line is the median, box extends from the 25th to the 75th percentile and whiskers denote minimum–maximum values. n = 8 biological replicates. **P = 0.0027, two-tailed, non-paired t-test.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4 and Supplementary Item 1 - Selective Enrichment of Corticospinal Neuronal Genes in Sampled BacTRAP Animals.
Supplementary Table 5
Complete dataset of significantly differentially expressed genes (FDR < 0.1) at day 10, 14 and 21 (logFC and p-values).
Rights and permissions
About this article
Cite this article
Poplawski, G.H.D., Kawaguchi, R., Van Niekerk, E. et al. Injured adult neurons regress to an embryonic transcriptional growth state. Nature 581, 77–82 (2020). https://doi.org/10.1038/s41586-020-2200-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2200-5
This article is cited by
-
Dual electrical stimulation at spinal-muscular interface reconstructs spinal sensorimotor circuits after spinal cord injury
Nature Communications (2024)
-
Transplantation of Heat-Shock Preconditioned Neural Stem/Progenitor Cells Combined with RGD-Functionalised Hydrogel Promotes Spinal Cord Functional Recovery in a Rat Hemi-Transection Model
Stem Cell Reviews and Reports (2024)
-
Phosphorylated proteomics-based analysis of the effects of semaglutide on hippocampi of high-fat diet-induced-obese mice
Diabetology & Metabolic Syndrome (2023)
-
Developmental stage of transplanted neural progenitor cells influences anatomical and functional outcomes after spinal cord injury in mice
Communications Biology (2023)
-
Spinal cord injury: molecular mechanisms and therapeutic interventions
Signal Transduction and Targeted Therapy (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.