Abstract
Separation of eukaryotic sister chromatids during the cell cycle is timed by the spindle assembly checkpoint (SAC) and ultimately triggered when separase cleaves cohesion-mediating cohesin1,2,3. Silencing of the SAC during metaphase activates the ubiquitin ligase APC/C (anaphase-promoting complex, also known as the cyclosome) and results in the proteasomal destruction of the separase inhibitor securin1. In the absence of securin, mammalian chromosomes still segregate on schedule, but it is unclear how separase is regulated under these conditions4,5. Here we show that human shugoshin 2 (SGO2), an essential protector of meiotic cohesin with unknown functions in the soma6,7, is turned into a separase inhibitor upon association with SAC-activated MAD2. SGO2–MAD2 can functionally replace securin and sequesters most separase in securin-knockout cells. Acute loss of securin and SGO2, but not of either protein individually, resulted in separase deregulation associated with premature cohesin cleavage and cytotoxicity. Similar to securin8,9, SGO2 is a competitive inhibitor that uses a pseudo-substrate sequence to block the active site of separase. APC/C-dependent ubiquitylation and action of the AAA-ATPase TRIP13 in conjunction with the MAD2-specific adaptor p31comet liberate separase from SGO2–MAD2 in vitro. The latter mechanism facilitates a considerable degree of sister chromatid separation in securin-knockout cells that lack APC/C activity. Thus, our results identify an unexpected function of SGO2 in mitotically dividing cells and a mechanism of separase regulation that is independent of securin but still supervised by the SAC.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All source data for this study are available online.
References
Musacchio, A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25, R1002–R1018 (2015).
Nasmyth, K. & Haering, C. H. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 (2009).
Uhlmann, F., Wernic, D., Poupart, M. A., Koonin, E. V. & Nasmyth, K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell 103, 375–386 (2000).
Mei, J., Huang, X. & Zhang, P. Securin is not required for cellular viability, but is required for normal growth of mouse embryonic fibroblasts. Curr Biol 11, 1197–1201 (2001).
Pfleghaar, K., Heubes, S., Cox, J., Stemmann, O. & Speicher, M. R. Securin is not required for chromosomal stability in human cells. PLoS Biol. 3, e416 (2005).
Llano, E. et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 22, 2400–2413 (2008).
Orth, M. et al. Shugoshin is a Mad1/Cdc20-like interactor of Mad2. EMBO J. 30, 2868–2880 (2011).
Boland, A. et al. Cryo-EM structure of a metazoan separase-securin complex at near-atomic resolution. Nat. Struct. Mol. Biol. 24, 414–418 (2017).
Lin, Z., Luo, X. & Yu, H. Structural basis of cohesin cleavage by separase. Nature 532, 131–134 (2016).
Wirth, K. G. et al. Separase: a universal trigger for sister chromatid disjunction but not chromosome cycle progression. J. Cell. Biol. 172, 847–860 (2006).
Zou, H., McGarry, T. J., Bernal, T. & Kirschner, M. W. Identification of a vertebrate sister-chromatid separation inhibitor involved in transformation and tumorigenesis. Science 285, 418–422 (1999).
Boos, D., Kuffer, C., Lenobel, R., Korner, R. & Stemmann, O. Phosphorylation-dependent binding of cyclin B1 to a Cdc6-like domain of human separase. J. Biol. Chem. 283, 816–823 (2008).
Hellmuth, S. et al. Positive and negative regulation of vertebrate separase by Cdk1-cyclin B1 may explain why securin is dispensable. J. Biol. Chem. 290, 8002–8010 (2015).
Huang, X. et al. Preimplantation mouse embryos depend on inhibitory phosphorylation of separase to prevent chromosome missegregation. Mol. Cell. Biol. 29, 1498–1505 (2009).
Huang, X. et al. Inhibitory phosphorylation of separase is essential for genome stability and viability of murine embryonic germ cells. PLoS Biol. 6, e15 (2008).
Huang, X., Hatcher, R., York, J. P. & Zhang, P. Securin and separase phosphorylation act redundantly to maintain sister chromatid cohesion in mammalian cells. Mol. Biol. Cell 16, 4725–4732 (2005).
Stemmann, O., Zou, H., Gerber, S. A., Gygi, S. P. & Kirschner, M. W. Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 (2001).
Li, M., York, J. P. & Zhang, P. Loss of Cdc20 causes a securin-dependent metaphase arrest in two-cell mouse embryos. Mol. Cell. Biol. 27, 3481–3488 (2007).
Hellmuth, S. et al. Human chromosome segregation involves multi-layered regulation of separase by the peptidyl-prolyl-isomerase Pin1. Mol. Cell 58, 495–506 (2015).
Gorr, I. H., Boos, D. & Stemmann, O. Mutual inhibition of separase and Cdk1 by two-step complex formation. Mol. Cell 19, 135–141 (2005).
Rodriguez-Bravo, V. et al. Nuclear pores protect genome integrity by assembling a premitotic and Mad1-dependent anaphase inhibitor. Cell 156, 1017–1031 (2014).
Holland, A. J. & Taylor, S. S. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J. Cell Sci. 119, 3325–3336 (2006).
Clift, D., Bizzari, F. & Marston, A. L. Shugoshin prevents cohesin cleavage by PP2A(Cdc55)-dependent inhibition of separase. Genes Dev. 23, 766–780 (2009).
Tsou, M. F. & Stearns, T. Mechanism limiting centrosome duplication to once per cell cycle. Nature 442, 947–951 (2006).
Kawashima, S. A., Yamagishi, Y., Honda, T., Ishiguro, K. & Watanabe, Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327, 172–177 (2010).
Hauf, S. et al. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 3, e69 (2005).
Eytan, E. et al. Disassembly of mitotic checkpoint complexes by the joint action of the AAA-ATPase TRIP13 and p31comet. Proc. Natl Acad. Sci. USA 111, 12019–12024 (2014).
Reddy, S. K., Rape, M., Margansky, W. A. & Kirschner, M. W. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446, 921–925 (2007).
Bakos, G. et al. An E2-ubiquitin thioester-driven approach to identify substrates modified with ubiquitin and ubiquitin-like molecules. Nat. Commun. 9, 4776 (2018).
Hellmuth, S., Böttger, F., Pan, C., Mann, M. & Stemmann, O. PP2A delays APC/C-dependent degradation of separase-associated but not free securin. EMBO J. 33, 1134–1147 (2014).
Gorr, I. H. et al. Essential CDK1-inhibitory role for separase during meiosis I in vertebrate oocytes. Nat. Cell Biol. 8, 1035–1037 (2006).
Hellmuth, S., Gutiérrez-Caballero, C., Llano, E., Pendás, A. M. & Stemmann, O. Local activation of mammalian separase in interphase promotes double-strand break repair and prevents oncogenic transformation. EMBO J. 37, e99184 (2018).
Schöckel, L., Möckel, M., Mayer, B., Boos, D. & Stemmann, O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 13, 966–972 (2011).
Murray, A. W. Cell cycle extracts. Methods Cell Biol. 36, 581–605 (1991).
McGuinness, B. E., Hirota, T., Kudo, N. R., Peters, J. M. & Nasmyth, K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3, e86 (2005).
Franken, N. A., Rodermond, H. M., Stap, J., Haveman, J. & van Bree, C. Clonogenic assay of cells in vitro. Nat. Protocols 1, 2315–2319 (2006).
Butt, T. R., Edavettal, S. C., Hall, J. P. & Mattern, M. R. SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 43, 1–9 (2005).
Holland, A. J., Bottger, F., Stemmann, O. & Taylor, S. S. Protein phosphatase 2A and separase form a complex regulated by separase autocleavage. J. Biol. Chem. 282, 24623–24632 (2007).
Schägger, H. Tricine-SDS-PAGE. Nat. Protocols 1, 16–22 (2006).
Acknowledgements
We thank S. Heidmann and P. Wolf for critical reading of the manuscript, and J. Hübner and M. Hermann for technical assistance. This work was supported by a grant (STE997/4-2) from the Deutsche Forschungsgemeinschaft (DFG) to O.S. and by MINECO (BFU2017-89408-R) and Junta de Castilla y Leon (CSI239P18). CIC-IBMCC is supported by the Programa de Apoyo a Planes Estratégicos de Investigación de Estructuras de Investigación de Excelencia cofunded by the Castilla–León autonomous government and the European Regional Development Fund (CLC–2017–01).
Author information
Authors and Affiliations
Contributions
L.G.-H. and A.M.P. first discovered the interaction between separase and SGO2. S.H. carried out all experiments except for the one shown in Extended Data Fig. 1a, which was conducted by L.G.-H. S.H. and O.S. co-designed the research and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Silke Hauf, Adele Marston and Hongtao Yu for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
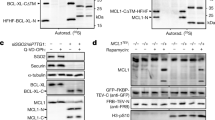
Extended Data Fig. 1 Mammalian SGO2 interacts with separase.
a, Hek293T cells were co-transfected with expression vectors for the following mouse proteins: GFP–separase, Flag–SGO2 and securin. Immunoprecipitation was carried out from transfected cells with either anti-Flag or anti-GFP antibodies and analysed by immunoblotting with the indicated antibodies. b, Mouse separase does not interact with human SGO2 and, therefore, cannot be used to study separase regulation in human cells. Securin-depleted Hek293T cells expressing GFP-tagged human separase(S1126A) or mouse separase(S1121A) were taxol-arrested and then subjected to IP–immunoblotting analyses using the indicated antibodies. c, SGO2 and MAD2 interact specifically with separase in untransformed cells. Taxol-arrested RPE1 cells were subjected to IP–immunoblotting analyses as indicated. d, Even in securin-expressing cells, a considerable fraction of separase is sequestered by SGO2. Taxol-arrested Hek293T, HCT116, and HeLa-K cells were subjected to IP–immunoblotting analyses using the indicated antibodies. ab 1, anti-DVPPRESHSHSDQSSKC (corresponding to amino acids 230–245 of human SGO2); ab 2, anti-KSEDLSSERTSRRRRC (corresponding to amino acids 1,234–1,249 of human SGO2); rb, rabbit; mo, mouse. Given below are the relative intensities (in per cent) of the separase signals (sum of full-length and N-terminal auto-cleavage fragment). Note the considerable co-depletion of separase upon SGO2 immunoprecipitation from HeLa-K (right).
Extended Data Fig. 2 Sgo1 rather than Sgo2 interacts with Mad2 and separase in Xenopus.
The indicated variants of in vitro-expressed X. laevis (X.l.) shugoshins and an excess of E.-coli-expressed Mad2 (to mimic SAC signalling) were incubated in anaphase egg extract (left). Following IP with anti-X.l. separase or mock-IgG from these mixtures, isolated proteins were detected by immunoblotting (middle and right). R170A, Mad2-binding-deficient Sgo1; ΔC, Sgo1-binding-deficient, C-terminally truncated Mad2.
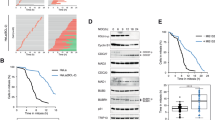
Extended Data Fig. 3 Lack of securin and SGO2 or MAD2 have synergistic cytotoxic effects.
a–c, Clonogenic assays with HeLa-K (a), PTTG1−/− (SECURIN−/−) and parental HCT116 cells (b), and Hek293T cells (c) transfected with the indicated siRNAs. Bars show percentages of colony numbers relative to the control of three independent experiments (dots).
Extended Data Fig. 4 Depletion of securin and SGO2, but not the individual knockdowns, results in impaired chromosome alignment, premature loss of chromosomal cohesin and unscheduled separase activity.
a, HeLa-K cells transfected with the indicated siRNAs and a histone H2B–eGFP expression plasmid were observed by video fluorescence microscopy to assess metaphase plate formation (bottom). Bars show mean of three independent experiments (dots) counting at least 50 mitotic cells each (top). Scale bar, 5 μm. b, Eight hours after release from thymidine arrest, HeLa-K cells transfected with the indicated siRNAs were pre-extracted, fixed, and examined by fluorescence microscopy for cohesin-negative early mitotic chromatin. Left, representative images; right, bars show mean of three independent experiments (dots) counting prophase nuclei that were still round but already stained positive for condensin (100 each). Scale bar, 5 μm. c, Hek293T cells transfected with the indicated siRNAs were supplemented with taxol (but not EGCG; compare Fig. 2b) and analysed by chromosome spreading. Bars show mean of three independent experiments (dots). d, Exemplary immunoblots of cells analysed in c and Fig. 2b. Star denotes nonspecific band. e, siRNA-transfected HeLa-K cells expressing a separase activity sensor (H2B-mCherry–RAD21107–268–eGFP) were released from a taxol arrest by addition of ZM-447439 at time zero and analysed by time-resolved immunoblotting.
Extended Data Fig. 5 Identification of SGO2-binding-deficient separase variants.
Taxol-arrested Hek293T cells expressing transgenic Myc-tagged wild-type separase (WT) or one of the indicated variants (V1–V5) were analysed by IP–immunoblotting using the indicated antibodies. The investigation of PP2A-binding-deficient variants was motivated by the notion that SGO2–MAD2, like PP2A, preferentially interacts with full-length rather than auto-cleaved separase38 (see, for example, Fig. 3c). V3 cannot bind PP2A but can bind SGO2–MAD2, indicating overlapping but not identical binding sites.
Extended Data Fig. 6 Depletion of securin and Sgo2 but not the individual knock-downs result in premature disengagement of centrioles.
HeLa-K cells transfected with the indicated siRNAs were released from a thymidine block and arrested in G2 phase with the CDK1 inhibitor RO-3306. Corresponding lysates were used for immunoblotting (bottom right) and centrosome isolation followed by immunofluorescence microscopy (top right, representative images) to assess the degree of centriole disengagement as revealed by two C-Nap1 foci. Left, bars show mean of three independent experiments (dots) counting 100 centrosomes each. Scale bar, 1 μm.
Extended Data Fig. 7 Mutually exclusive binding of SGO2 to MAD2 or phosphorylated histone H2A.
a–d, Pre-charging of SGO2 with phosphorylated histone H2A blocks subsequent MAD2 binding. a, Experimental scheme for experiments shown in b–d. Beads loaded with the indicated Flag–SGO2 variants were consecutively incubated first with phosphorylated or unphosphorylated H2A or H3 peptide (input 1) and then with wild-type or, where indicated, C-terminally truncated (ΔC) MAD2 (input 2). Following washing (supernatant 2), bound proteins were visualized by immunoblotting. b, Usage of free H2A peptides. c, Usage of ovalbumin-coupled H2A peptides to facilitate their detection by standard glycine-SDS–PAGE and immunoblotting. d, Phosphorylated histone H3 peptide does not bind to SGO2. Free H2A and H3 peptides, used to interrogate immobilized SGO2, were separated by Tricine-SDS–PAGE39 and analysed by Coomassie staining and immunoblotting. e, Pre-charging of SGO2 with MAD2 blocks subsequent phospho-H2A binding. Experimental scheme (top) and corresponding immunoblotting analysis (bottom). H2A peptides were coupled to ovalbumin for ease of detection. f, Cartoon illustrating that SGO2 can bind to H2A phosphorylated on Thr121 or to MAD2 but not both at the same time.
Extended Data Fig. 8 Mapping separase-binding sites on X. laevis Sgo1.
a, Covering amino acids 200–300 of X. laevis Sgo1 with polyclonal antibodies impairs its binding to separase. Region-specific polyclonal X. laevis Sgo1 antibodies A–D were characterized (left) and added to in vitro-expressed X. laevis Sgo1 and E.-coli-expressed MAD2 (prey) as indicated. Immobilized X. laevis separase isolated by immunoprecipitation from anaphase egg extracts was then added as bait to these mixtures. Separase beads were washed and finally probed for associated proteins by immunoblotting. Mock, unspecific IgG; asterisk, unspecific band. b, c, Identification of two sites within X. laevis Sgo1 that are relevant for separase binding. Different in vitro translated (IVT) X. laevis Sgo1 fragments and variants thereof were combined with wild-type MAD2 or MAD2-ΔC as indicated (prey). Separase beads as in a were combined with these mixtures, washed and analysed for associated proteins by immunoblotting. d–f, X. laevis Sgo1(143–145A) and Sgo1(254–256A) (red) show compromised separase binding but retain MAD2 binding. d, The indicated full-length X. laevis Sgo1 variants were assessed for MAD2-dependent separase binding as in b, c. e, The indicated X. laevis Sgo1 fragments and variants thereof were combined with wild-type MAD2 or MAD2-ΔC, immunoprecipitated via their Myc-tag and assessed for MAD2 binding by immunoblotting. f, Summary of the mapping experiments. MIM, Mad2-interaction motif; n.d., not determined.
Extended Data Fig. 9 X. laevis Sgo1 and human SGO2 share the same order and spacing of separase- and MAD2-binding sites.
a, A point mutation turns X. laevis Sgo1 into a Mad2-dependent separase substrate. 35S-labelled X. laevis Sgo1 variants were incubated with wild-type Mad2 or Mad2-ΔC before being assayed for in vitro-cleavage by X. laevis separase, which had been immunoprecipitated from anaphase egg extracts. Gels were blotted onto membranes, which were cut and subjected to immunoblotting (top) before reassembly and autoradiography (bottom). b, Human (H.s.) SGO2(127–129A) and SGO2(239–242A) show compromised separase binding but retain MAD2 binding. The indicated full-length SGO2 variants were immunoprecipitated via their Flag-tags from transfected, taxol-arrested Hek293T cells and assessed by immunoblotting for binding of MAD2 and separase. See also illustration at bottom. c, Cartoon comparing the arrangement of functional domains and alignment of pseudo-substrate and Mad2-binding sites of X. laevis Sgo1 and human SGO2. The separase interaction site around position 144 of X. laevis Sgo1 and around position 128 of human SGO2 was omitted for clarity.
Extended Data Fig. 10 In vitro disassembly of separase–SGO2–MAD2 by APC/CCDC20-dependent ubiquitylation.
Immobilized separase–SGO2–MAD2 complex isolated as in Fig. 4a was incubated with ATP, ubiquitin, E1, UBE2S, wild-type or dominant-negative (DN) UBE2C and APC/CCDC20 (or reference buffer). After removal of the supernatant and washing, the beads were incubated with 35S-RAD21 before being analysed by autoradiography and immunoblotting. The autoradiograph shows the relevant upper and lower parts of the same gel. Ubiquitylation of purified securin served as a positive control.
Supplementary information
Supplementary Data
This file contains gel source data scans.
Rights and permissions
About this article
Cite this article
Hellmuth, S., Gómez-H, L., Pendás, A.M. et al. Securin-independent regulation of separase by checkpoint-induced shugoshin–MAD2. Nature 580, 536–541 (2020). https://doi.org/10.1038/s41586-020-2182-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2182-3
This article is cited by
-
Genome-wide CRISPR screen identifies ESPL1 limits the response of gastric cancer cells to apatinib
Cancer Cell International (2024)
-
Genome control by SMC complexes
Nature Reviews Molecular Cell Biology (2023)
-
SGOL2 is a novel prognostic marker and fosters disease progression via a MAD2-mediated pathway in hepatocellular carcinoma
Biomarker Research (2022)
-
Clinical significance of FBXW7 loss of function in human cancers
Molecular Cancer (2022)
-
Structural basis of human separase regulation by securin and CDK1–cyclin B1
Nature (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.