Abstract
Radiologic screening of high-risk adults reduces lung-cancer-related mortality1,2; however, a small minority of eligible individuals undergo such screening in the United States3,4. The availability of blood-based tests could increase screening uptake. Here we introduce improvements to cancer personalized profiling by deep sequencing (CAPP-Seq)5, a method for the analysis of circulating tumour DNA (ctDNA), to better facilitate screening applications. We show that, although levels are very low in early-stage lung cancers, ctDNA is present prior to treatment in most patients and its presence is strongly prognostic. We also find that the majority of somatic mutations in the cell-free DNA (cfDNA) of patients with lung cancer and of risk-matched controls reflect clonal haematopoiesis and are non-recurrent. Compared with tumour-derived mutations, clonal haematopoiesis mutations occur on longer cfDNA fragments and lack mutational signatures that are associated with tobacco smoking. Integrating these findings with other molecular features, we develop and prospectively validate a machine-learning method termed ‘lung cancer likelihood in plasma’ (Lung-CLiP), which can robustly discriminate early-stage lung cancer patients from risk-matched controls. This approach achieves performance similar to that of tumour-informed ctDNA detection and enables tuning of assay specificity in order to facilitate distinct clinical applications. Our findings establish the potential of cfDNA for lung cancer screening and highlight the importance of risk-matching cases and controls in cfDNA-based screening studies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Anonymized clinical and demographic data on the lung cancer cases and non-cancer controls considered in this study, as well as cfDNA metrics, cfDNA and WBC somatic mutation data, Lung-CLiP scores, and other relevant data are provided in the Supplementary Tables. The detailed patient-level genomic features used as input for the Lung-CLiP model (including genome-wide somatic copy number alteration data and somatic mutation genotyping data with all the associated features considered in the Lung-CLiP model), along with code for the Lung-CLiP classification model, the in silico simulation of the CAPP-Seq molecular biology workflow, and the modified dNdScv R functions38 (accounting for the fraction of a given gene covered by our sequencing panel) can be found at http://clip.stanford.edu. This website provides users with the code and data used for the training and validation of the Lung-CLiP model and the in silico simulation of the CAPP-Seq molecular biology workflow, allowing for reproduction of our results and figures. Owing to restrictions related to dissemination of germline sequence information included in the informed consent forms used to enrol study subjects, we are unable to provide access to raw sequencing data. Reasonable requests for additional data will be reviewed by the senior authors to determine whether they can be fulfilled in accordance with these privacy restrictions. Requests for additional materials related to this work should be directed to M.D.
References
The National Lung Screening Trial Research Team. Results of initial low-dose computed tomographic screening for lung cancer. N. Engl. J. Med. 368, 1980–1991 (2013).
de Koning, H. J. et al. Reduced lung-cancer mortality with volume CT screening in a randomized trial. N. Engl. J. Med. 382, 503–513(2020).
Jemal, A. & Fedewa, S. A. Lung cancer screening with low-dose computed tomography in the United States—2010 to 2015. JAMA Oncol. 3, 1278–1281 (2017).
Doria-Rose, V. P. et al. Use of lung cancer screening tests in the United States: results from the 2010 National Health Interview Survey. Cancer Epidemiol. Biomarkers Prev. 21, 1049–1059 (2012).
Newman, A. M. et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 34, 547–555 (2016).
Moyer, V. A. Screening for lung cancer: U.S. Preventive Services Task Force recommendation statement. Ann. Intern. Med. 160, 330–338 (2014).
Pinsky, P. F. et al. Performance of Lung-RADS in the National Lung Screening Trial. Ann. Inter. Med. 162, 485 (2015).
Chaudhuri, A. A. et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 7, 1394–1403 (2017).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017); corrigendum 554, 264 (2018).
Jiang, P. et al. Lengthening and shortening of plasma DNA in hepatocellular carcinoma patients. Proc. Natl Acad. Sci. USA 112, E1317–E1325 (2015).
Mouliere, F. et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 10, eaat4921 (2018).
Travis, W. D. et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 6, 244–285 (2011).
Moding, E. J. et al. Circulating tumor DNA dynamics predict benefit from consolidation immunotherapy in locally advanced non-small-cell lung cancer. Nat. Cancer 1, 176–183 (2020).
Steensma, D. P. et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 126, 9–16 (2015).
Lui, Y. Y. N. et al. Predominant hematopoietic origin of cell-free DNA in plasma and serum after sex-mismatched bone marrow transplantation. Clin. Chem. 48, 421–427 (2002).
Liu, J. et al. Biological background of the genomic variations of cf-DNA in healthy individuals. Ann. Oncol. 30, 1–7 (2018).
Razavi, P. et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 25, 1928–1937 (2019).
Ptashkin, R. N. et al. Prevalence of clonal hematopoiesis mutations in tumor-only clinical genomic profiling of solid tumors. JAMA Oncol. 4, 1589–1593 (2018).
The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525 (2012).
The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma Nature 511, 543–550 (2014).
Alexandrov, L. B. et al. Signatures of mutational processes in human cancer. Nature 500, 415–421 (2013).
Hainaut, P. & Pfeifer, G. P. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb. Perspect. Med. 6, a026179 (2016).
Shen, S. Y. et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 563, 579–583 (2018).
Cohen, J. D. et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 359, 926–930 (2018).
Phallen, J. et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9, eaan2415 (2017).
Cristiano, S. et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 570, 385–389 (2019).
Simon, R. Roadmap for developing and validating therapeutically relevant genomic classifiers. J. Clin. Oncol. 23, 7332–7341 (2005).
Ma, J., Ward, E. M., Smith, R. & Jemal, A. Annual number of lung cancer deaths potentially avertable by screening in the United States. Cancer 119, 1381–1385 (2013).
Kurtz, D. M. et al. Dynamic risk profiling using serial tumor biomarkers for personalized outcome prediction. Cell 178, 699–713.e19 (2019).
Chen, S. et al. AfterQC: automatic filtering, trimming, error removing and quality control for fastq data. BMC Bioinformatics 18, 80 (2017).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Karczewski, K. J. et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. Preprint at https://www.biorxiv.org/content/10.1101/531210v2 (2019).
Koboldt, D. C. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22, 568–576 (2012).
Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31, 213–219 (2013).
Saunders, C. T. et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 28, 1811–1817 (2012).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385.e18 (2018).
Martincorena, I. et al. Universal patterns of selection in cancer and somatic tissues. Cell 171, 1029–1041.e21 (2017).
Rosenthal, R., McGranahan, N., Herrero, J., Taylor, B. S. & Swanton, C. deconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016).
Hindson, B. J. et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83, 8604–8610 (2011).
Mermel, C. H. et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Acknowledgements
We thank E. Kool for advice relating to ROS scavengers and E. Edell and A. Bungum from the Mayo Clinic Lung Tumor Specimen Registry for their assistance with sample collection. This work was supported by the National Cancer Institute (R01CA188298 and R01CA233975 to M.D. and A.A.A., 1-K08-CA241076-01 to D.M.K., R25CA180993 and T32-CA 121940 to M.S.E., U01 CA196405 to P.P.M., training grant T32 CA009302 to E.G.H., M.C.N. and D.A., and K12CA090628 and P30 CA015083-44S1 to A.S.M.), the US National Institutes of Health Director’s New Innovator Award Program (1-DP2-CA186569 to M.D.), the US National Institutes of Health, the Virginia and D.K. Ludwig Fund for Cancer Research (M.D. and A.A.A.), the CRK Faculty Scholar Fund (M.D.), the Bakewell Foundation (M.D. and A.A.A.), the Damon Runyon Cancer Research Foundation (PST#09-16 to D.M.K.), the Tobacco-Related Disease Research Program Predoctoral Fellowship (T30DT0806 to E.G.H.), the Blavatnik Family Fellowship (E.G.H.), the American Cancer Society (134031-PF-19-164-01-TBG to B.Y.N.), the SDW/DT and Shanahan Family Foundations (A.A.A.), Stand Up To Cancer (M.D., A.A.A., D.A.H. and L.V.S.), and the NSF Graduate Research Fellowship (DGE-114747 to J.J.C., DGE-1656518 to D.A.). A.A.A. is a Scholar of The Leukemia & Lymphoma Society.
Author information
Authors and Affiliations
Contributions
J.J.C., E.G.H., D.M.K., M.S.E., A.A.A. and M.D. developed the concept, designed the experiments and analysed the data. J.J.C., E.G.H., D.M.K., M.S.E., A.A.A. and M.D. wrote the manuscript. J.J.C. and D.M.K. developed the in silico simulation of the CAPP-Seq workflow and the FLEX adaptors with input from M.D. and A.A.A. J.J.C. and M.S.E. developed the machine learning module of the Lung-CLiP model with input from E.G.H. and D.M.K. J.J.C. performed molecular biology experiments related to improving the CAPP-Seq workflow. J.J.C. and E.G.H. performed molecular biology related to profiling clinical specimens with assistance from E.J.M., B.Y.N., A.A.C., A.B.H., T.D.A., Y.-J.J., M.C.N. and D.A. Bioinformatics analyses were performed by J.J.C., E.G.H., D.M.K., M.S.E., H.S., J.S.-M., B.C., C.L.L., M.C.J., M.C.N., L.C.T.K. and R.T. Patient specimens were provided by P.P.M., A.S.M., J.J., S.H.L., C.L.C., R.B., J.W.N., H.A.W., B.W.L., N.S.L., M.F.B., J.B.S., S.S.G., V.S.N., D.A.H., L.V.S. and M.D. A.N.L. performed radiologic analyses. R.F.B., C.H.Y., R.B.K., E.L.C., D.J.M., P.P.M., H.Z.R., A.S.M., C.L.C., R.B., G.J.B., K.C.J., R.B.W., C.A.K. and V.S.N. organised patient enrolment, sample collection and clinical data curation. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
D.M.K. reports paid consultancy from Roche Molecular Diagnostics. A.A.C. reports speaker honoraria and travel support from Roche Sequencing Solutions, Varian Medical Systems, and Foundation Medicine, a research grant from Roche Sequencing Solutions, and has served as a paid consultant for Fenix Group International. A.S.M. reports advisory for AbbVie, Genentech, and Bristol-Myers Squibb (honoraria paid to institution) and research funding from Novartis and Verily. J.J. is now employed by Celgene. S.H.L. reports paid advisory from AstraZeneca, speaker honoraria from Varian Medical Systems and research funding from BeyondSpring Pharmaceuticals Inc., Hitachi Chemical Diagnostics, Genentech, and New River Labs. S.S.G. reports paid consultancy from AbbVie, Ceremark, CytomX Therapeutics Inc., GPV, Life Molecular Imaging, Nusano, Spectrum Dynamics, and TPG, and ownership interest in Akrotome Imaging Inc., Cellsight Technologies, CytomX Therapeutics Inc., Earli Inc., Endra Inc., MagArray Inc., Nines, Nodus Therapeutics, Nusano, RefleXion Medical Inc., SiteOne Therapeutics Inc., Spectrum Dynamics, Vave Health, and Vor Biopharma. J.W.N. reports paid consultancy from AstraZeneca, Genentech, Roche, Exelixis, Jounce Therapeutics, Takeda Pharmaceuticals, Eli Lilly and Company, and Calithera Biosciences, and research funding from Genentech, Roche, Merck, Novartis, Boehringer Ingelheim, Exelixis, Nektar Therapeutics, Takeda Pharmaceuticals, Adaptimmune and GSK. H.A.W. reports paid advisory from AstraZeneca, Xcovery, Janssen, and Mirati, unpaid advisory from Merck, Takeda, Genentech, Roche, and Cellworks, and research funding from ACEA Biosciences, Arrys Therapeutics, AstraZeneca/Medimmune, BMS, Celgene, Clovis Oncology, Exelixis, Genentech/Roche, Gilead, Lilly, Merck, Novartis, Pfizer, Pharmacyclics, and Xcovery. A.A.A. reports ownership interest in CiberMed and FortySeven Inc., patent filings related to cancer biomarkers, and paid consultancy from Genentech, Roche, Chugai, Gilead, and Celgene. M.D. reports research funding from Varian Medical Systems and Illumina, ownership interest in CiberMed, patent filings related to cancer biomarkers, and paid consultancy from Roche, AstraZeneca, RefleXion and BioNTech. The remaining authors declare no potential conflicts of interest.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Development and experimental validation of an in silico simulation of the CAPP-Seq molecular biology workflow.
a, The fraction of original unique (blue line) and duplex (green line) cfDNA molecules (unique depth; right axis) and total molecules including PCR duplicates (nondeduped depth; left axis) at each step in the CAPP-Seq molecular biology workflow were tracked using an in silico model based on random binomial sampling. In this model only on-target molecules are considered, with both individual DNA strands from original DNA duplexes tracked. Two simulations are shown, with 8.3% (top) and 100% (bottom) of amplified sequencing library input into the hybridization reaction for target enrichment. Additional details on the model are provided in the Supplementary Methods. b, c, Empirical validation of simulation models. Comparison of median unique de-duplicated (that is, ‘deduped’) (b) and duplex (c) depths recovered by sequencing following the input of different fractions of sequencing library into the hybrid capture reaction. A total of 32 ng of cfDNA from each of four healthy adults was used as the input in each condition and each sample was downsampled to 100 million sequencing reads before barcode-deduplication to facilitate comparison. Comparisons were performed with a paired two-sided t-test. d, e, Comparison of deduped (d) and duplex (e) sequencing depths achieved following the input of 8.3% (n = 138 cfDNA samples) compared to ≥25% (n = 145 cfDNA samples) of each sequencing library into the hybrid capture reaction. All samples had 32 ng of cfDNA as the input to the library preparation and were downsampled to 25 million reads before barcode-deduplication to facilitate comparison. In box plots the centre line denotes the median, the box contains the interquartile range, and the whiskers denote the extrema that are no more than 1.5 × IQR from the edge of the box (Tukey style). f, g, Comparison of deduped (f) and duplex (g) sequencing depths predicted by the model to that observed experimentally when 8.3% or 100% of a sequencing library is input into the hybrid capture reaction. A range of capture efficiencies (7.5–75% hybrid capture efficiency) were considered in the simulation, in which the confidence envelope denotes the resultant range of model predictions. The experimental data depicted in b, c (n = 4 cfDNA samples per capture condition) was downsampled before barcode deduplication to enable comparisons across different sequencing read yields (x axis). Dots denote the median and error bars denote the minimum and maximum.
Extended Data Fig. 2 The ROS scavenger hypotaurine reduces oxidative damage arising in vitro.
a, Diagram illustrating the chemical mechanism by which carcinogens in cigarette smoke in vivo (top) or ROS in vitro (bottom) cause damage to DNA leading to the generation of 8-oxoguanine, which subsequently results in the generation of G>T transversions. b, Diagram illustrating the proposed mechanism by which the addition of a ROS scavenger reduces oxidative-damage-derived G>T artefacts in vitro. c, Comparison of the distribution of base substitutions in healthy control cfDNA samples (n = 12 individuals) captured with and without the ROS scavenger hypotaurine present in the hybrid capture reaction. The number of errors that are G>T transversions was compared using a paired two-sided t-test (P < 1 × 10−8). d, e, Aggregate selector-wide nondeduped (d) and deduped (e) background error rates summarizing the results in c. Grouped comparisons were performed with a paired two-sided t-test. f, Comparison of selector-wide deduped background error rates and base substitution distributions across two cohorts of healthy controls, in which cfDNA samples were profiled with (present; bottom, n = 104) or without (absent; top, n = 69) the ROS scavenger hypotaurine in the hybrid capture reaction. g, Aggregate selector-wide error rates summarizing the results from f. In box plots the centre line denotes the median, the box contains the interquartile range, and the whiskers denote the extrema that are no more than 1.5 × IQR from the edge of the box (Tukey style).
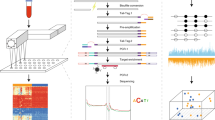
Extended Data Fig. 3 Rationale for and overview of dual-index duplex adaptors with error-correcting barcodes (FLEX adaptors).
a, An excess of molecular barcodes (that is, unique identifier or UIDs) differing by 1 bp in cfDNA molecules with the same the start and end positions indicates that sequencing errors in UIDs can create erroneous UID families. Depicted are the expected and observed distributions of barcode Hamming edit distances (UID edit distance) when comparing UIDs from different groups of barcode-deduped (that is, unique) cfDNA molecules sequenced using our previously described tandem adaptors5. Tandem adaptors utilize random 4-mer UIDs, resulting in 256 distinct UIDs that cannot be error corrected. The theoretical distribution of UID edit distances across all 256 UIDs is shown in orange (that is, the fraction of UIDs that differ from one another by 1, 2, 3, and 4 bp). The green, red and blue bars represent the distribution of UID edit distances observed in healthy control cfDNA samples sequenced with tandem adaptors (n = 24 individuals). Green indicates randomly sampled UIDs, blue indicates UIDs from cfDNA molecules with different genomic start and end positions, and red indicates cfDNA molecules that share the same start and end positions. UIDs differing by only one base are significantly overrepresented when comparing cfDNA molecules with the same start and end position (red bars) to each of the other UID distributions, suggesting that 1-bp errors are erroneously creating new UID families. Group comparisons were performed with a paired two-sided t-test, except when comparing to the theoretical distribution, for which an unpaired two-sided t-test was used (P < 1 × 10−8). Bars denote the mean and error bars denote the standard error of the mean. b, Schematic overview of custom FLEX sequencing adaptors, enabling independent tailoring of UID diversity and multiplexing capacity. Shown is an initial DNA molecule to which partial Y adaptors containing duplex UIDs are ligated (1–2). Next, the two molecules derived after one round of grafting PCR—which adds the first of two sample barcodes—are shown (3). This is followed by additional rounds of grafting PCR, which add the second sample barcode and continue to amplify the library (4). After grafting PCR, a magnetic bead cleanup is performed (not shown) and is followed by universal PCR (5), after which final sequencing libraries compatible with Illumina sequencers are shown (6). Dual-index sample barcodes types are indicated in yellow (index 1 or i7) and orange (index 2 or i5) and UIDs are indicated by purple and green blocks. c, Diagram depicting a detailed view of the partial Y adaptors used for initial ligation to cfDNA. The adaptors contain a 1-bp offset that is indicated in green, followed by a 6-bp error-correcting UID indicated in purple (Hamming edit distances ≥ 3), followed by 0–3 ‘stagger’ bases indicated in red, followed by a 3′ T-overhang for ligation. The 0–3-bp stagger bases increase sequence complexity early in the sequencing reads to obviate the need for PhiX (used for spectral diversity). Additional details on the FLEX adaptors are provided in the Supplementary Methods.
Extended Data Fig. 4 Study and cohort overview.
a, Overview of the study. b, Clinical and demographic information pertaining to the NSCLC patient cohorts and the non-cancer control cohorts considered in this study. For categorical variables, the count is provided with the percentage of the cohort in parentheses. For continuous variables, the median value is provided with the range of values in parentheses. NOS, not otherwise specified. aAJCC v7 staging. bLow-risk controls were considered for feature discovery and clonal haematopoiesis analysis only and were not used for Lung-CLiP model training. cSex was compared with a two-sided Fisher’s exact test and continuous variables (age and pack-years) were compared with an unpaired two-sided t-test. dLung-CLiP patients with NSCLC and risk-matched controls were compared.
Extended Data Fig. 5 Biological determinants of tumour-informed ctDNA detection.
a, Association between tumour-informed ctDNA detection and the number of mutations tracked using the population-based lung-cancer-focused CAPP-Seq panel. All patients were considered and binned by the number of mutations identified in matched tumour biopsy samples. b, Association between the number of mutations identified in matched tumour samples and tumour-informed ctDNA detection using the population-based lung-cancer-focused CAPP-Seq panel. c, ctDNA detection statistics in 17 patients with early-stage NSCLC profiled with both the population-based lung-cancer-focused CAPP-Seq panel (left) and customized capture panels designed using tumour exome sequencing data (right). Whereas ctDNA in all 17 patients was undetectable using the population-based method, it was detected in 10 (59%) patients using customized panels. For patients with detectable ctDNA, the mean VAF observed across all tracked mutations is depicted (blue circles). For samples without detectable ctDNA, the corresponding patient-specific analytical LOD is shown (open circles). LOD was determined on the basis of the binomial distribution, number of mutations tracked and the median unique molecular depth in the sample. When calculating the LOD in samples sequenced with the population-based panel, deduped depth was considered. When calculating the LOD in samples sequenced with customized panels, duplex depth was considered if this gave an LOD below the deduped error rate. In both scenarios, if the LOD was less than the background error rate for the cfDNA molecule type being considered (either deduped or duplex), the background error rate was used. d, Comparison of the patient-specific analytical LOD in patients with and without detectable ctDNA using tumour-informed CAPP-Seq. LOD was determined as in c and the LOD in samples sequenced with the population-based lung-cancer-focused CAPP-Seq panel only (n = 68) and samples sequenced with customized capture panels designed using tumour exome sequencing data (n = 17) are displayed. e, Detection of clonal and subclonal SNVs in cfDNA. The fraction of all clonal and subclonal SNVs detected in plasma are depicted in pie charts (two-sided Fisher’s exact test, P = 0.039) and the VAFs of clonal and subclonal SNVs detectable in plasma are compared using violin plots in which horizontal dashed lines depict the median and interquartile range. All mutations identified using the population-based lung-cancer-focused CAPP-Seq panel are considered. f, The fraction of all mutant and wild-type cfDNA molecules (defined as in Fig. 1d) with fragment sizes falling within the size windows found to be ctDNA-enriched in Fig. 1e. g, Violin plot displaying the enrichment of SNV VAFs following in silico size selection for the cfDNA fragment sizes found to be ctDNA-enriched in Fig. 1e. Enrichment is defined as the ratio of the SNV VAF after size selection to that observed before size selection. All mutations identified in matched tumor samples and detectable in plasma before size selection (n = 323 mutations) were considered. In the box plot, the centre line denotes the median, the box contains the interquartile range, and the whiskers denote the extrema that are no more than 1.5 × IQR from the edge of the box (Tukey style). h, Comparison of SNV VAFs before and after size selection. The dot plot displays the VAF of SNVs in plasma before and after size selection. The bar plot depicts the fraction of SNVs for which the VAF increased, decreased or became undetectable after size selection. All mutations identified in matched tumor samples and detectable in plasma before size selection were considered. i, Comparison of SNV VAFs before size selection in SNVs for which the VAF increased, decreased, or became undetectable after size selection. All mutations identified in matched tumor samples and detectable in plasma before size selection were considered. j, Tumour-informed ctDNA detection rates before and after size selection in patients sequenced with the population-based lung-cancer-focused CAPP-Seq panel (n = 85 patients) and customized capture panels designed using tumour exome sequencing data (n = 17 patients).
Extended Data Fig. 6 Clinical correlates of tumour-informed ctDNA detection.
a, Relationship between MTV measured by PET-CT and pretreatment ctDNA concentration measured in haploid genome equivalents per ml plasma (hGE ml−1). All patients with detectable ctDNA and MTV measurements available were considered (n = 46). The comparison was performed by Spearman correlation. b, Comparison of MTV in patients with and without detectable ctDNA. All patients with MTV measurements (n = 81) were considered. c, Multiple variable linear regression was performed to associate the predictor variables (MTV, histology and stage) with mean ctDNA VAF. For patients without detectable ctDNA, a VAF of 0.0001% was used. All patients with MTV measurements (n = 81) were considered. Additional details are provided in the Methods. d, Comparison of pretreatment ctDNA levels in patients with adenocarcinoma histology and varying amounts of GGO on pretreatment CT scans. The brackets above the plot depict the comparison (Fisher’s exact test) between ctDNA detection in patients with <25% GGO (24/48 patients with ctDNA detected) and patients with ≥25% GGO (2/13 patients with ctDNA detected). Top, representative CT scans of tumors with different amounts of GGO with the lesions outlined. All patients with adenocarcinoma histology and pretreatment CT scans available were considered (n = 61). e, ctDNA detection rates in all patients (n = 82, blue bars) and in only those with adenocarcinoma histology (n = 61, grey bars) with tumours that do or do not have evidence of necrosis on pretreatment CT scans. Top, representative CT scans of tumors that do (right) and do not (left) have evidence of necrosis; lesions are outlined and regions of necrosis are indicated with an arrow. Detection rates were compared by Fisher’s exact test. All patients with pretreatment CT scans available were considered (n = 82).
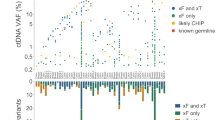
Extended Data Fig. 7 Pretreatment ctDNA burden is prognostic in early-stage NSCLC.
a–d, Kaplan–Meier analysis for recurrence-free survival (a, b) and freedom from metastasis (c, d) stratified by pretreatment ctDNA level in all patients with stage I–III disease (a, c, n = 85) and in patients with stage I disease only (b, d, n = 48). The median ctDNA level across the cohort (0.0031%) was used to stratify patients into ctDNA-high and ctDNA-low groups. P values were calculated using the log-rank test. e, Table summarizing the results of univariable and multiple variable Cox proportional hazards models. MTV measured by PET-CT and ctDNA measurements (mean SNV VAF) were log transformed. Significant P values (<0.05) are shown in bold. For univariable analysis of ctDNA level and stage, all patients (n = 85) were considered. For the univariable analysis of MTV, and for each multiple variable analysis, only patients with MTV measurements available (n = 81) were considered. Univariable and multiple variable P values were assessed using the log-likelihood test. f, Example data from patients with stage I adenocarcinoma. Left, data from two patients with high pretreatment ctDNA levels who developed distant metastases after surgery. Right, data from two patients with undetectable ctDNA who achieved long term remission after surgery.
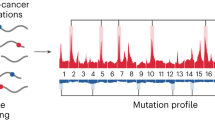
Extended Data Fig. 8 Biological features of cfDNA mutations reflecting clonal haematopoiesis.
a, Flow chart depicting the fraction of WBC+ and WBC− cfDNA mutations affecting canonical clonal haematopoiesis genes in patients with NSCLC and controls. WBC+ cfDNA mutations present at ≥1% VAF in matched leukocytes more frequently affect canonical clonal haematopoiesis genes than those present at levels below 1% (51/64 versus 223/460 WBC+ cfDNA mutations present at ≥1% versus <1% VAF in matched leukocytes affect canonical CH genes, respectively; P = 1.9 × 10−6, Fisher’s exact test). Only mutations identified de novo in the cfDNA for which presence in the matched WBCs could be confidently assessed are considered (Methods). b, The percentage of mutations genotyped de novo from WBC DNA at VAFs of <2% and ≥2% affecting canonical clonal haematopoiesis genes in patients and controls (all patients and controls are considered). The comparison was performed by Fisher’s exact test. c, The percentage of controls (left) and patients with NSCLC (right) with one or more mutations in the ten genes that most frequently contained WBC+ cfDNA mutations. Patients with NSCLC and controls with only WBC+ mutations, only WBC− mutations, or both WBC+ and WBC− mutations in a gene are depicted in red, grey and pink, respectively. The numbers next to each bar represent the percentage of all cfDNA mutations in that gene that are WBC+ in patients with NSCLC (right) or controls (left). Patients with NSCLC had significantly more WBC− cfDNA mutations in TP53 than controls (19/32 and 0/4 in patients and controls, respectively. *P = 0.04, Fisher’s exact test). d, Mutation frequency by gene for WBC+ cfDNA mutations observed across all patients with NSCLC (n = 104) and controls (n = 98). The y axis depicts the percentage of the combined cohort with WBC+ cfDNA mutations affecting a given gene. All genes with mutations in four or more individuals in the combined cohort are depicted. e, Scatter plot comparing the VAFs of WBC+ cfDNA mutations across multiple time points in patients with NSCLC (left panel, n = 54 mutations, n = 8 individuals) and controls (right panel, n = 12 mutations, n = 6 individuals). The statistical comparison was performed by Pearson correlation on mutations detected at both time points. f, Positive selection analysis was carried out on all synonymous and nonsynonymous WBC+ (n = 693 mutations, red) and WBC− (n = 526 mutations, grey) cfDNA mutations observed in patients with NSCLC and controls using the dNdScv R package with a modification to account for the fraction of a given gene covered by our sequencing panel. The x axis indicates the dNdScv adjusted P value (Q value) for all substitution types. Genes were considered under positive selection if the Q value was less than 0.05. All genes meeting this threshold are displayed. Additional details are provided in the Methods. g, Distribution of WBC+ and WBC− cfDNA mutations across the p53 protein in patients with NSCLC and controls. h, Short fragment enrichment of WBC+ and WBC− cfDNA mutations in patients with NSCLC and controls, defined as the fold change in VAF for a given mutation after in silico size selection for the cfDNA fragment sizes found to be ctDNA-enriched in Fig. 1e. The centre line denotes the median, the box contains the interquartile range, and the whiskers denote the 10th and 90th percentile values.
Extended Data Fig. 9 Feature importance and performance of Lung-CLiP.
a, Biological and technical parameters specific to each individual variant used as features in a dedicated logistic regression ‘SNV model’. The feature names are depicted on the y axis, and the negative log10 of the P value derived from comparing all post-filtered SNVs in patients with NSCLC (n = 574 mutations from n = 104 individuals) with those in risk-matched controls (n = 64 mutations from n = 56 individuals) in a univariable linear model in the training set is shown on the x axis. All features with a P value of less than 0.01 are shown, P values were calculated using an unpaired two-sided t-test. Additional information about each feature is provided in the Supplementary Methods. b, Receiver operating characteristic (ROC) curves for the Lung-CLiP model depicting performance stratified by tumour stage in the training set (n = 104 patients with NSCLC and n = 56 risk-matched controls). c, Spectrum of clinicopathologic correlates and selected features observed across the 46 patients with early-stage NSCLC and 48 risk-matched controls undergoing annual lung cancer screening in a prospectively enrolled independent validation cohort. d, Receiver operating characteristic curves for the Lung-CLiP model depicting performance stratified by tumour stage in the validation set (n = 46 patients with NSCLC and n = 48 risk-matched controls). e, Comparison of the specificity observed in the validation cohort at different thresholds defined in the training cohort. Dots denote the median specificity across 1,000 bootstrap resamplings and error bars depict the interquartile range. Statistical comparison was performed by Pearson correlation on the non-bootstrapped data. f–i, Comparison of metabolic tumour volume (f), cfDNA input to library preparation (g), plasma volume used (h) and unique sequencing depth (i) in patients with NSCLC correctly classified at 98% specificity (positive) to those in patients that were incorrectly classified (negative). All patients with NSCLC in the training and validation cohorts were considered (n = 103 patients with metabolic tumour volume measurements in f and n = 150 patients in g–i). In box plots, the centre line denotes the median, the box contains the interquartile range, and the whiskers denote the extrema that are no more than 1.5 × IQR from the edge of the box (Tukey style).
Extended Data Fig. 10 Technical reproducibility and benchmarking of CAPP-Seq and the Lung-CLiP model.
a–j, Blood was drawn from each of three healthy donors into two Streck tubes and two K2EDTA tubes and processed using the protocols used in our study. cfDNA extraction and library preparation were performed as described in the Methods with 25 ng of cfDNA input for each sample. Sequencing and data processing were performed as described in the Methods and each sample was downsampled to 80 million reads before barcode-deduplication to facilitate comparison. a, The Lung-CLiP model was trained on the 104 patients with NSCLC and 56 risk-matched controls in the training cohort and applied to the cfDNA samples extracted from plasma drawn into Streck and K2EDTA tubes. The fraction of donors classified as negative by Lung-CLiP at the 98% (blue bars) and 80% (red bars) specificity thresholds defined in the training data are depicted. b–h, Comparison of median cfDNA fragment size (b), cfDNA concentration in ng ml−1 (c), deduped depth (d), duplex depth (e) and error metrics (f–h) in cfDNA samples extracted from plasma drawn into the two tube types. cfDNA samples from the same donor are connected with dashed lines, comparisons were performed using a paired two-sided t-test. i, Comparison of the fragment size distribution of cfDNA samples extracted into the two tube types. j, Genotyping was performed as described in the Methods on cfDNA samples extracted from plasma drawn into the two tube types from the three donors. Donor 1 and donor 3 each had one mutation identified in cfDNA that was present in samples extracted from plasma drawn into both tube types and was also present in matched WBCs (WBC+). Donor 2 had no mutations identified in cfDNA samples extracted from plasma drawn into either tube type. k, Orthogonal validation of WBC+ cfDNA mutations (n = 15) using ddPCR. Comparison of the VAF of WBC+ cfDNA mutations as measured by CAPP-Seq (x axis) and ddPCR (y axis). ddPCR was performed in triplicate (technical replicates) on cfDNA (left) or WBC DNA (right) sequencing libraries. All 15 mutations (100%) were validated by ddPCR in both the cfDNA and WBC compartments. Triangles represent recurrent ‘hotspot’ mutations in canonical clonal haematopoiesis genes and squares represent private mutations in non-clonal haematopoiesis genes. The points denote the median and error bars denote the minimum and maximum. Statistical comparison was performed by Pearson correlation. l–n, Tumour-informed ctDNA levels in patients with NSCLC, with and without adjustments for copy-number state and clonality of tumour mutations. l, VAFs of individual mutations (n = 323) observed in cfDNA with different SNV VAF adjustment strategies. Comparisons were performed using a paired two-sided t-test. m, The mean cfDNA VAF across all tracked mutations tracked in patients with detectable ctDNA (n = 48) with the different adjustment strategies. Comparisons were performed using a paired two-sided t-test. n, The same data as in m separated by stage. In box plots the centre line denotes the median, the box contains the interquartile range, and the whiskers denote the extrema that are no more than 1.5 × IQR from the edge of the box (Tukey style). In l–n, copy number and clonality adjustment was performed as described in the Supplementary Methods.
Supplementary information
Supplementary Information
Supplementary Methods: This file contains additional methodological details relating to 1) the simulation of the CAPP-Seq molecular biology workflow, 2) the FLEX sequencing adapters, 3) detection of genome-wide copy number variation from targeted sequencing data, 4) clonality and copy number state adjustment for tumor-informed ctDNA detection, and 5) the Lung-CLIP model.
Supplementary Information
Supplementary Note: This file contains a description of the motivation for this study.
Supplementary Tables
This excel file contains Supplementary Tables 1-11. These tables contain a variety of supporting information related to this study (e.g. demographic and clinical information on participants, ctDNA detection metrics, cfDNA and WBC mutation data, Lung-CLiP scores…etc). Descriptions of the contents of each table are provided.
Rights and permissions
About this article
Cite this article
Chabon, J.J., Hamilton, E.G., Kurtz, D.M. et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 580, 245–251 (2020). https://doi.org/10.1038/s41586-020-2140-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2140-0
This article is cited by
-
Early Detection of Lung Cancer with Low-Dose CT Scan Using Artificial Intelligence: A Comprehensive Survey
SN Computer Science (2024)
-
Finite element method and hybrid deep learning approaches: high-accuracy lung cancer detection model
Multiscale and Multidisciplinary Modeling, Experiments and Design (2024)
-
Plasma cell-free DNA as a sensitive biomarker for multi-cancer detection and immunotherapy outcomes prediction
Journal of Cancer Research and Clinical Oncology (2024)
-
Distinct Hodgkin lymphoma subtypes defined by noninvasive genomic profiling
Nature (2024)
-
Prediction of plasma ctDNA fraction and prognostic implications of liquid biopsy in advanced prostate cancer
Nature Communications (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.