Abstract
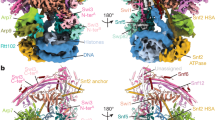
Chromatin-remodelling complexes of the SWI/SNF family function in the formation of nucleosome-depleted, transcriptionally active promoter regions (NDRs)1,2. In the yeast Saccharomyces cerevisiae, the essential SWI/SNF complex RSC3 contains 16 subunits, including the ATP-dependent DNA translocase Sth14,5. RSC removes nucleosomes from promoter regions6,7 and positions the specialized +1 and −1 nucleosomes that flank NDRs8,9. Here we present the cryo-electron microscopy structure of RSC in complex with a nucleosome substrate. The structure reveals that RSC forms five protein modules and suggests key features of the remodelling mechanism. The body module serves as a scaffold for the four flexible modules that we call DNA-interacting, ATPase, arm and actin-related protein (ARP) modules. The DNA-interacting module binds extra-nucleosomal DNA and is involved in the recognition of promoter DNA elements8,10,11 that influence RSC functionality12. The ATPase and arm modules sandwich the nucleosome disc with the Snf2 ATP-coupling (SnAC) domain and the finger helix, respectively. The translocase motor of the ATPase module engages with the edge of the nucleosome at superhelical location +2. The mobile ARP module may modulate translocase–nucleosome interactions to regulate RSC activity5. The RSC–nucleosome structure provides a basis for understanding NDR formation and the structure and function of human SWI/SNF complexes that are frequently mutated in cancer13.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lorch, Y. & Kornberg, R. D. Chromatin-remodeling for transcription. Q. Rev. Biophys. 50, e5 (2017).
Clapier, C. R., Iwasa, J., Cairns, B. R. & Peterson, C. L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422 (2017).
Cairns, B. R. et al. RSC, an essential, abundant chromatin-remodeling complex. Cell 87, 1249–1260 (1996).
Saha, A., Wittmeyer, J. & Cairns, B. R. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16, 2120–2134 (2002).
Clapier, C. R. et al. Regulation of DNA translocation efficiency within the chromatin remodeler RSC/Sth1 potentiates nucleosome sliding and ejection. Mol. Cell 62, 453–461 (2016).
Krietenstein, N. et al. Genomic nucleosome organization reconstituted with pure proteins. Cell 167, 709–712 (2016).
Klein-Brill, A., Joseph-Strauss, D., Appleboim, A. & Friedman, N. Dynamics of chromatin and transcription during transient depletion of the RSC chromatin remodeling complex. Cell Rep. 26, 279–292 (2019).
Kubik, S. et al. Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol. Cell 60, 422–434 (2015).
Ramachandran, S., Zentner, G. E. & Henikoff, S. Asymmetric nucleosomes flank promoters in the budding yeast genome. Genome Res. 25, 381–390 (2015).
Badis, G. et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32, 878–887 (2008).
Lorch, Y., Maier-Davis, B. & Kornberg, R. D. Role of DNA sequence in chromatin remodeling and the formation of nucleosome-free regions. Genes Dev. 28, 2492–2497 (2014).
Kubik, S. et al. Sequence-directed action of RSC remodeler and general regulatory factors modulates +1 nucleosome position to facilitate transcription. Mol Cell 71, 89–102 (2018).
Pulice, J. L. & Kadoch, C. Composition and function of mammalian SWI/SNF chromatin remodeling complexes in human disease. Cold Spring Harb. Symp. Quant. Biol. 81, 53–60 (2016).
Chambers, A. L., Pearl, L. H., Oliver, A. W. & Downs, J. A. The BAH domain of Rsc2 is a histone H3 binding domain. Nucleic Acids Res. 41, 9168–9182 (2013).
Kasten, M. et al. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23, 1348–1359 (2004).
VanDemark, A. P. et al. Autoregulation of the Rsc4 tandem bromodomain by Gcn5 acetylation. Mol. Cell 27, 817–828 (2007).
Li, M. et al. Mechanism of DNA translocation underlying chromatin remodelling by Snf2. Nature 567, 409–413 (2019).
Saha, A., Wittmeyer, J. & Cairns, B. R. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 12, 747–755 (2005).
Schubert, H. L. et al. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc. Natl Acad. Sci. USA 110, 3345–3350 (2013).
Szerlong, H. et al. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat. Struct. Mol. Biol. 15, 469–476 (2008).
Sen, P. et al. The SnAC domain of SWI/SNF is a histone anchor required for remodeling. Mol. Cell. Biol. 33, 360–370 (2013).
Lorch, Y., Maier-Davis, B. & Kornberg, R. D. Histone acetylation inhibits RSC and stabilizes the +1 nucleosome. Mol. Cell. 72, 594–600 (2018).
Cakiroglu, A. et al. Genome-wide reconstitution of chromatin transactions reveals that RSC preferentially disrupts H2AZ-containing nucleosomes. Genome Res. 29, 988–998 (2019).
Suto, R. K., Clarkson, M. J., Tremethick, D. J. & Luger, K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7, 1121–1124 (2000).
Materne, P. et al. Histone H2B ubiquitylation represses gametogenesis by opposing RSC-dependent chromatin remodeling at the ste11 master regulator locus. eLife 5, e13500 (2016).
Brahma, S. & Henikoff, S. RSC-associated subnucleosomes define MNase-sensitive promoters in yeast. Mol. Cell 73, 238–249 (2019).
Brogaard, K., Xi, L., Wang, J. P. & Widom, J. A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501 (2012).
Dechassa, M. L. et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol. Cell 38, 590–602 (2010).
Patel, A. B. et al. Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement. eLife 8, e54449 (2019).
Ye, Y. et al. Structure of the RSC complex bound to the nucleosome. Science 366, 838–843 (2019).
Han, Y., Reyes, A. A., Malik, S. & He, Y. Cryo-electron microscopy structure of a nucleosome-bound SWI/SNF chromatin remodeling complex. Preprint at https://www.bioRxiv.org/content/10.1101/805184v1 (2019).
Cairns, B. R. et al. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4, 715–723 (1999).
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 (1999).
Lorch, Y. & Kornberg, R. D. Isolation and assay of the RSC chromatin-remodeling complex from Saccharomyces cerevisiae. Methods Enzymol. 377, 316–322 (2003).
Luger, K., Rechsteiner, T. J. & Richmond, T. J. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol. Biol. 119, 1–16 (1999).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2003).
Maskell, D. P. et al. Structural basis for retroviral integration into nucleosomes. Nature 523, 366–369 (2015).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Kastner, B. et al. GraFix: sample preparation for single-particle electron cryomicroscopy. Nat. Methods 5, 53–55 (2008).
Stark, H. GraFix: stabilization of fragile macromolecular complexes for single particle cryo-EM. Methods Enzymol. 481, 109–126 (2010).
Tegunov, D. & Cramer, P. Real-time cryo-EM data pre-processing with Warp. Nat. Methods 16, 1146–1152 (2019).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Asturias, F. J., Chung, W. H., Kornberg, R. D. & Lorch, Y. Structural analysis of the RSC chromatin-remodeling complex. Proc. Natl Acad. Sci. USA 99, 13477–13480 (2002).
Chaban, Y. et al. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat. Struct. Mol. Biol. 15, 1272–1277 (2008).
Leschziner, A. E. et al. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc. Natl Acad. Sci. USA 104, 4913–4918 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Kidmose, R. T. et al. Namdinator—automatic molecular dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ 6, 526–531 (2019).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Song, Y. et al. High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013).
Raman, S. et al. Structure prediction for CASP8 with all-atom refinement using Rosetta. Proteins 77 (Suppl 9), 89–99 (2009).
van Dijk, M. & Bonvin, A. M. 3D-DART: a DNA structure modelling server. Nucleic Acids Res. 37, W235–W239 (2009).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 (2018).
Bienert, S. et al. The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res. 45, D313–D319 (2017).
Charlop-Powers, Z., Zeng, L., Zhang, Q. & Zhou, M. M. Structural insights into selective histone H3 recognition by the human Polybromo bromodomain 2. Cell Res. 20, 529–538 (2010).
Da, G. et al. Structure and function of the SWIRM domain, a conserved protein module found in chromatin regulatory complexes. Proc. Natl Acad. Sci. USA 103, 2057–2062 (2006).
Legge, G. B. et al. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 343, 1081–1093 (2004).
Reichen, C. et al. Structures of designed armadillo-repeat proteins show propagation of inter-repeat interface effects. Acta Crystallogr. D 72, 168–175 (2016).
Grimm, M., Zimniak, T., Kahraman, A. & Herzog, F. xVis: a web server for the schematic visualization and interpretation of crosslink-derived spatial restraints. Nucleic Acids Res. 43, W362–W369 (2015).
Zimmermann, L. et al. A completely reimplemented mpi bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
Buchan, D. W. A. & Jones, D. T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019).
Jones, D. T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 (1999).
Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Schrodinger, LLC. The PyMOL Molecular Graphics System version 1.8 (2015).
Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Treich, I., Ho, L. & Carlson, M. Direct interaction between Rsc6 and Rsc8/Swh3,two proteins that are conserved in SWI/SNF-related complexes. Nucleic Acids Res. 26, 3739–3745 (1998).
Taneda, T. & Kikuchi, A. Genetic analysis of RSC58, which encodes a component of a yeast chromatin remodeling complex, and interacts with the transcription factor Swi6. Mol. Genet. Genomics 271, 479–489 (2004).
Angus-Hill, M. L. et al. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7, 741–751 (2001).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Liu, Y., Schmidt, B. & Maskell, D. L. MSAProbs: multiple sequence alignment based on pair hidden Markov models and partition function posterior probabilities. Bioinformatics 26, 1958–1964 (2010).
Bond, C. S. & Schüttelkopf, A. W. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D 65, 510–512 (2009).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Combe, C. W., Fischer, L. & Rappsilber, J. xiNET: cross-link network maps with residue resolution. Mol. Cell. Proteomics 14, 1137–1147 (2015).
Yan, Z. et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19, 1662–1667 (2005).
Xue, Y. et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl Acad. Sci. USA 97, 13015–13020 (2000).
Nicolas, R. H. & Goodwin, G. H. Molecular cloning of polybromo, a nuclear protein containing multiple domains including five bromodomains, a truncated HMG-box, and two repeats of a novel domain. Gene 175, 233–240 (1996).
Wang, W. et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl Acad. Sci. USA 95, 492–498 (1998).
Satterwhite, E. et al. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood 98, 3413–3420 (2001).
Sandhya, S., Maulik, A., Giri, M. & Singh, M. Domain architecture of BAF250a reveals the ARID and ARM-repeat domains with implication in function and assembly of the BAF remodeling complex. PLoS One 13, e0205267 (2018).
Worden, E. J., Hoffmann, N. A., Hicks, C. W. & Wolberger, C. Mechanism of cross-talk between H2B ubiquitination and H3 methylation by Dot1L. Cell 176, 1490–1501 (2019).
Eustermann, S. et al. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386–390 (2018).
Farnung, L., Vos, S. M., Wigge, C. & Cramer, P. Nucleosome–Chd1 structure and implications for chromatin remodelling. Nature 550, 539–542 (2017).
Willhoft, O. et al. Structure and dynamics of the yeast SWR1-nucleosome complex. Science 362, eaat7716 (2018).
Ayala, R. et al. Structure and regulation of the human INO80-nucleosome complex. Nature 556, 391–395 (2018).
Sundaramoorthy, R. et al. Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome. eLife 7, e35720 (2018).
Yan, L., Wu, H., Li, X., Gao, N. & Chen, Z. Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling. Nat. Struct. Mol. Biol. 26, 258–266 (2019).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Acknowledgements
We thank current and former members of the Cramer Laboratory, including S. Osman, G. Kokic, P. Seweryn, S. Schilbach, S. Neyer and H. Hillen. F.R.W. was supported by a Boehringer Ingelheim Fonds PhD fellowship. H.U. was supported by the Deutsche Forschungsgemeinschaft (SFB860). P.C. was supported by the Deutsche Forschungsgemeinschaft (SFB860, SPP1935, EXC 2067/1-390729940), the European Research Council Advanced Investigator Grant TRANSREGULON (grant agreement no. 693023), and the Volkswagen Foundation.
Author information
Authors and Affiliations
Contributions
F.R.W. carried out all experiments and data analysis unless stated otherwise. C.D. assisted with data collection and model building. A.S. and H.U. carried out cross-linking and mass spectrometry analysis. H.W. helped with nucleosome biochemistry. D.T. helped with cryo-EM data processing. P.C. designed and supervised the project. F.W. and P.C. wrote the manuscript with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Blaine Bartholomew and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Preparation and characterization of RSC–nucleosome complex.
a, Preparation of endogenous Rsc2-containing isoform of the RSC complex from S. cerevisiae. Analysis of purified RSC by size-exclusion chromatography and SDS–PAGE showed high purity and homogeneity with stoichiometric subunits as assessable by Coomassie stain. Subunit identity was confirmed by mass spectrometry. The table shows the expected molecular weights of the RSC subunits. For gel source data, see Supplementary Fig. 1. b, Assembly of the RSC–nucleosome complex. SDS–PAGE analysis of fractions 7–20 of a 10–25% sucrose-gradient ultracentrifugation. Complex formation was successful as demonstrated by the comigration of histones with the RSC complex. The unbound over-stoichiometric nucleosomes only migrated to fractions 7 and 8 (black arrow). Fraction 16 in the presence of cross-linker was used for cryo-EM grid preparation (dashed box). c, Location of cross-linking sites mapped onto the structure. BS3 cross-links that appeared at least in triplicates were mapped onto the RSC-nucleosome structure. Lysine residues involved in the cross-linking network are shown as blue spheres and cross-linked residues are connected with lines indicating permitted (blue) and non-permitted (red) cross-linking distances. Of the mapped cross-links, 87.5% are within the permitted cross-linking distance, which was set to 30 Å. The remaining 12.5% of non-permitted cross-links probably reflect ambiguity caused by the presence of two identical Rsc8 subunits in the structure as well as flexibility of the complex in buffer or arise from technical errors. d, The cross-linking network between subunits of the RSC–nucleosome complex. Subunits are coloured as in Fig. 1. Cross-links with a score above 2.5 are shown. A comprehensive list of cross-links is presented in Supplementary Data 1. Cross-linking mass-spectrometry experiments were performed in duplicates with similar results.
Extended Data Fig. 2 Cryo-EM analysis of the RSC–nucleosome complex.
a, Representative cryo-EM micrograph of the RSC–nucleosome complex shows homogeneously distributed individual particles. b–d, Two-dimensional class averages of the RSC–nucleosome complex (b), the ATPase–nucleosome subcomplex (c) and the nucleosome subcomplex (d). e, FSC plots reveal the overall resolutions of the cryo-EM reconstructions. f, Cryo-EM processing workflow for the reconstructions of the RSC–nucleosome complex, the ATPase–nucleosome subcomplex and the nucleosome subcomplex. Particle distribution after 3D classifications is indicated below the corresponding map. The final maps are shown in colour. The masks used for focused classifications and refinements are colour coded corresponding to the final maps they were used for. Views are generally rotated by 180° with respect to Fig. 1c, left. g, Local resolution estimation of the combined ATPase–nucleosome map as implemented in RELION42. We note that the resolution of the peripheral area with the ATPase module is overestimated. h–j, Angular distribution plot for all particles contributing to the final reconstructions of the RSC–nucleosome complex (h), the ATPase–nucleosome subcomplex (i) and the nucleosome subcomplex (j).
Extended Data Fig. 3 Cryo-EM analysis of the free RSC complex.
a, Representative cryo-EM micrograph of the free RSC complex shows homogeneously distributed individual particles. b, Two-dimensional class averages of the free RSC complex. c, Cryo-EM processing workflow for the reconstruction of the free RSC complex. Particle distribution after 3D classifications is indicated below the corresponding map. The final maps after focused 3D refinement and masks are depicted in colour. Views are generally rotated by 180° with respect to Fig. 1c, right. d, Angular distribution plot for all particles contributing to the final reconstruction of the free RSC complex. e, Two views of the combined RSC core map coloured according to the local resolution as implemented in RELION42. f, FSC plots of the maps used for model building of the RSC core complex.
Extended Data Fig. 4 Cryo-EM densities for selected RSC regions.
a–c, Examples of map quality. a, Close-up of the Rsc4 β-sheet shows clear separation of individual strands. b, The high quality of the map for the ZZ zinc-finger of Rsc8 allowed backbone tracing and placement of side chains as well as for the zinc ion. c, Coiled-coil helices of the two Rsc8 subunits with density for one helix. d, View along the exit DNA in the direction of the nucleosome showing the lowpass-filtered maps for the modules ATPase, ARP, DIM, arm and body, and the nucleosome. At the site where the H2A C-terminal tail protrudes from the nucleosome near Sfh1, there is low-resolution density connecting the arm module and the nucleosome. Density bridging form the ARP module to the exit DNA close to the H3 histone tail can be observed. e, Density representing the finger helix (green) at the acidic patch of the nucleosome (indicated by H2A in yellow). Side-chain density is visible for conserved arginine residues. f, Interaction of RSC with the nucleosome is sterically impaired by the flexibly bound ubiquitin moiety at H2B lysine 123 (K120 in human). The Sfh1 finger helix and the ubiquitin moiety (ubiquitylated nucleosome PDB code 6NOG)81 overlap after superposition of nucleosomes.
Extended Data Fig. 5 Structure of RSC body and arm modules, cancer mutations and remodeller families.
a, Cartoon representation of RSC core viewed as in Fig. 1. Important structural elements are labelled. b, Conservation between SWI/SNF complexes RSC (yeast) and PBAF (human). Residues that are identical (blue) or conserved (light blue) in human PBAF highlighted on the RSC structure (grey). Purple spheres depict identical residues that show missense mutations in various cancers. c, Comparison of overall structure of RSC with complexes of INO80 (yeast INO80)82 and CHD (yeast CHD1)83 families. ATPase motor domains are shown in orange, DNA is shown in blue. With regard to the INO80 family, the ATPase of the SWR1 complex also binds SHL +2 (ref. 84), whereas the ATPase of the INO80 complex binds SHL −6 (refs. 82,85). The ARP module of INO80 contacts exit DNA, which is not the case in RSC. The INO80 complex also contacts both faces of the histone octamer82, resembling the sandwiching interactions made by RSC on a topological level. With respect to the CHD family, the ATPase motor of yeast Chd1 also binds SHL +2, but its DNA-binding region engages with exit DNA near the nucleosome, leading to a different DNA trajectory83,86. With respect to the ISWI family, the ATPase motor binds SHL +2 (ref. 87), but other interactions have not been structurally resolved (not shown).
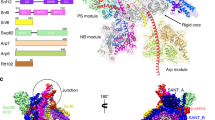
Extended Data Fig. 6 Course of polypeptide chains of architectural subunits Sth1, Rsc8 and Rsc58 and ATPase–nucleosome interactions.
a, Back view of RSC. The Sth1 subunit of RSC starts with its N terminus in the body module and tracks through it, turning around with a contact helix and loop. Forming the central helix I, the hook and the central helix II it folds back and forth tightly interweaving the body module before it exits with its HSA region through the ARP module to build the ATPase module. b, RSC with the domains of the two Rsc8 subunits highlighted in blue. Both Rsc8 start N-terminal with their SWIRM domains in the arm module where they support the two repeat domains of Sfh1 in a similar manner. They then follow distinct paths through the arm towards the body module where they contribute with both their SANT and ZZ zinc-finger domains. Here the two domains of each subunit form different contacts with various interactions partners and whereas one ZZ zinc-finger domain is tightly packed at the body and DNA-interaction module interface, the other seems to extend from the body, presumably as additional interaction surface. Both Rsc8 subunits unite again with their C-terminal long helices in a coiled-coil fold on the opposite side of the body module. c, Rsc58 N-terminal bromodomain attaches to the top of the body module. Then, Rsc58 follows an interwound path through the body module via the central and connector loop. It turns back, docking to the body with a three-helix bundle and stabilizing the module with its C-terminal end. d, Contacts of Sth1 ATPase motor (orange) with the nucleosome. View as in Fig. 1c, left, but rotated by 45° around a horizontal axis. Arrows indicate directionality of DNA translocation.
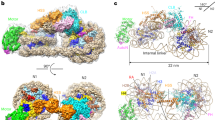
Extended Data Fig. 7 DNA recognition and NDR formation.
a, Space-filling RSC–nucleosome structure with DIM (green) and SnAC (orange) densities. View on the top as in Fig. 1c, left, but rotated by 90° around the vertical and horizontal axis. Arrows indicate directionality of DNA translocation. Number of upstream DNA base pairs relative to SHL −7 is provided. b, Schematic of a promoter before (top) and after (bottom) RSC remodelling shows NDR formation by sliding the flanking –1 and +1 nucleosomes away from the NDR centre. Arrows indicate the transcription start site.
Extended Data Fig. 8 Sequence alignments for the Sth1 ATPase domain and HSA region.
a, Sequence alignment of the S. cerevisiae Sth1 ATPase domain to the homologous Snf2 ATPase domain of the same organism. Secondary structure elements are represented in orange according to the cryo-EM structure of the Snf2 ATPase (PDB entry 5Z3U)17. Residues modelled in the Snf2 structure are topped by a black line with helical regions shown as cylinders and sheet regions as arrows. The Sth1 residues modelled in this work are indicated with a black dashed line below. ATPase motifs are underlined. Invariant residues are coloured in dark blue and conserved residues in light blue. The alignment was generated with MSAProbs71 within the MPI Bioinformatics Toolkit60 and visualized using ESPript88. b, Sequence alignment of the HSA regions from S. cerevisiae homologues Sth1 and Snf2. Illustration and generation of the alignment as in a.
Supplementary information
Supplementary Figure
Supplementary Figure 1 | Uncropped gel images with size marker. Uncropped gel images for Extended Data Figs. 1a and 1b. * Sample from another unrelated experiment, not from the size exclusion chromatography run.
Supplementary Table
Supplementary Table 1 | RSC subunit modelling. Modelling details for the RSC complex. Density that could not be assigned to a subunit was modelled with a poly alanine backbone in chain X. The domains of the two Rsc8 subunits could not be connected. Therefore, they are clustered by proximity and combined in one chain (L) spaced by a 443 amino acid number offset.
Supplementary Table
Supplementary Table 2 | Cryo-EM data collection, refinement and validation statistics. * Dataset from two collections were combined. Datasets from three collections were combined, † not tilted, ‡ 25° tilt. # Reported value is based on visual inspection, because the software-generated values were overestimated. § Map7: chains A, B, C, D, E, F, G, H, I (2-124), J (24-146), K (390-408) and S (393-1006); Map8: chains K (151-389), L, M, N, O, P, Q, R, S (48-297) and X.
Supplementary Data
Supplementary Data 1 | RSC-nucleosome BS3 crosslinks. List of intra- and inter-subunit lysine-lysine crosslinks as identified by LC-MS analyses and subsequent database search using pLink 2. The respective scores of cross-link identification are listed as well as the number of CSMs (cross-linked spectra matches). The numbering for the histone H3 is shifted because the N-terminal PEP sequence was omitted for purification (instead of MPEPAKSAP... it is MAKSAP...). The numbering for the Rsc2 protein includes the remains of C-terminal TAP-tag after TEV cleavage.
Video 1
Overview of RSC structure. The video shows the structure of RSC rotating around a vertical axis. It first depicts the low pass-filtered cryo-EM map, showing the five lobes of RSC and the nucleosome with exit DNA extending from it. It then shows the high-resolution cryo-EM maps for RSC modules, and finally the structural model as a ribbon representation with subunits in different colours (colour code as in Fig. 1).
Rights and permissions
About this article
Cite this article
Wagner, F.R., Dienemann, C., Wang, H. et al. Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome. Nature 579, 448–451 (2020). https://doi.org/10.1038/s41586-020-2088-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2088-0
This article is cited by
-
Asymmetric nucleosome PARylation at DNA breaks mediates directional nucleosome sliding by ALC1
Nature Communications (2024)
-
Structure of the ISW1a complex bound to the dinucleosome
Nature Structural & Molecular Biology (2024)
-
Energy-driven genome regulation by ATP-dependent chromatin remodellers
Nature Reviews Molecular Cell Biology (2024)
-
Structure of nucleosome-bound human PBAF complex
Nature Communications (2022)
-
SMARCC2 mediates the regulation of DKK1 by the transcription factor EGR1 through chromatin remodeling to reduce the proliferative capacity of glioblastoma
Cell Death & Disease (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.