Abstract
The production of pore-forming toxins that disrupt the plasma membrane of host cells is a common virulence strategy for bacterial pathogens such as methicillin-resistant Staphylococcus aureus (MRSA)1,2,3. It is unclear, however, whether host species possess innate immune mechanisms that can neutralize pore-forming toxins during infection. We previously showed that the autophagy protein ATG16L1 is necessary for protection against MRSA strains encoding α-toxin4—a pore-forming toxin that binds the metalloprotease ADAM10 on the surface of a broad range of target cells and tissues2,5,6. Autophagy typically involves the targeting of cytosolic material to the lysosome for degradation. Here we demonstrate that ATG16L1 and other ATG proteins mediate protection against α-toxin through the release of ADAM10 on exosomes—extracellular vesicles of endosomal origin. Bacterial DNA and CpG DNA induce the secretion of ADAM10-bearing exosomes from human cells as well as in mice. Transferred exosomes protect host cells in vitro by serving as scavengers that can bind multiple toxins, and improve the survival of mice infected with MRSA in vivo. These findings indicate that ATG proteins mediate a previously unknown form of defence in response to infection, facilitating the release of exosomes that serve as decoys for bacterially produced toxins.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The datasets generated herein are available from the corresponding authors upon request. Western blot gel source data can be found in Supplementary Fig. 1. All identified proteins from mass-spectrometry experiments and thier accession identification codes are listed in Supplementary Tables 1–3. Source Data for Fig. 4 and Extended Data Fig. 6 are available with the paper.
References
Dal Peraro, M. & van der Goot, F. G. Pore-forming toxins: ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 (2016).
Seilie, E. S. & Bubeck Wardenburg, J. Staphylococcus aureus pore-forming toxins: the interface of pathogen and host complexity. Semin. Cell Dev. Biol. 72, 101–116 (2017).
Spaan, A. N., van Strijp, J. A. G. & Torres, V. J. Leukocidins: staphylococcal bi-component pore-forming toxins find their receptors. Nat. Rev. Microbiol. 15, 435–447 (2017).
Maurer, K. et al. Autophagy mediates tolerance to Staphylococcus aureus alpha-toxin. Cell Host Microbe 17, 429–440 (2015).
Inoshima, I. et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat. Med. 17, 1310–1314 (2011).
Becker, R. E., Berube, B. J., Sampedro, G. R., DeDent, A. C. & Bubeck Wardenburg, J. Tissue-specific patterning of host innate immune responses by Staphylococcus aureus α-toxin. J. Innate Immun. 6, 619–631 (2014).
Matsuzawa-Ishimoto, Y., Hwang, S. & Cadwell, K. Autophagy and inflammation. Annu. Rev. Immunol. 36, 73–101 (2018).
Presley, J. F., Mayor, S., McGraw, T. E., Dunn, K. W. & Maxfield, F. R. Bafilomycin A1 treatment retards transferrin receptor recycling more than bulk membrane recycling. 272, 13929–13936 (1997).
Johnson, L. S., Dunn, K. W., Pytowski, B. & McGraw, T. E. Endosome acidification and receptor trafficking: bafilomycin A1 slows receptor externalization by a mechanism involving the receptor's internalization motif. 4, 1251–1266 (1993).
Cadwell, K. & Debnath, J. Beyond self-eating: the control of nonautophagic functions and signaling pathways by autophagy-related proteins. J. Cell Biol. 217, 813–822 (2018).
Stoeck, A. et al. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 393, 609–618 (2006).
Shimoda, M. et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nat. Cell Biol. 16, 889–901 (2014).
Ejlerskov, P. et al. Tubulin polymerization-promoting protein (TPPP/p25α) promotes unconventional secretion of α-synuclein through exophagy by impairing autophagosome-lysosome fusion. J. Biol. Chem. 288, 17313–17335 (2013).
Guo, H. et al. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev. Cell 43, 716–730 (2017).
Miao, Y., Li, G., Zhang, X., Xu, H. & Abraham, S. N. A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell 161, 1306–1319 (2015).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Kimura, T. et al. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 36, 42–60 (2017).
Trajkovic, K. et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247 (2008).
Cho, Y. E. et al. Increased liver-specific proteins in circulating extracellular vesicles as potential biomarkers for drug- and alcohol-induced liver injury. PLoS One 12, e0172463 (2017).
Alonzo, F., III et al. CCR5 is a receptor for Staphylococcus aureus leukotoxin ED. Nature 493, 51–55 (2013).
Mitamura, T., Higashiyama, S., Taniguchi, N., Klagsbrun, M. & Mekada, E. Diphtheria toxin binds to the epidermal growth factor (EGF)-like domain of human heparin-binding EGF-like growth factor/diphtheria toxin receptor and inhibits specifically its mitogenic activity. J. Biol. Chem. 270, 1015–1019 (1995).
Montecalvo, A. et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766 (2012).
Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Invest. 126, 1208–1215 (2016).
Henry, B. D. et al. Engineered liposomes sequester bacterial exotoxins and protect from severe invasive infections in mice. Nat. Biotechnol. 33, 81–88 (2015).
Tan, J. M. J. et al. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat. Microbiol. 3, 1472–1485 (2018).
Matsuzawa-Ishimoto, Y. et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J. Exp. Med. 214, 3687–3705 (2017).
Murrow, L., Malhotra, R. & Debnath, J. ATG12–ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 17, 300–310 (2015).
Guo, H. et al. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Dev. Cell 43, 716–730 (2017).
Hubbard-Lucey, V. M. et al. Autophagy gene Atg16L1 prevents lethal T cell alloreactivity mediated by dendritic cells. Immunity 41, 579–591 (2014).
Marchiando, A. M. et al. A deficiency in the autophagy gene Atg16L1 enhances resistance to enteric bacterial infection. Cell Host Microbe 14, 216–224 (2013).
DuMont, A. L. et al. Staphylococcus aureus LukAB cytotoxin kills human neutrophils by targeting the CD11b subunit of the integrin Mac-1. Proc. Natl Acad. Sci. USA 110, 10794–10799 (2013).
van der Vlist, E. J., Nolte-’t Hoen, E. N., Stoorvogel, W., Arkesteijn, G. J. & Wauben, M. H. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat. Protocols 7, 1311–1326 (2012).
Théry, C., Amigorena, S., Raposo, G. & Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 30, 3.22.21–3.22.29 (2006).
Acknowledgements
We thank members of the Cadwell and Torres laboratory for constructive comments over the years, and particularly I. Irnov and E. Anderson for providing purified S. aureus DNA and assisting with the LukED protection experiments, respectively. This work was supported in part by US National Institute of Health (NIH) grants R01 AI121244 (to K.C. and V.J.T.); R01 HL123340, R01 DK093668, R01 DK103788, R01 AI130945 and R01 HL125816 (to K.C.); R01 AI099394 and R01 AI105129 (to V.J.T.); T32 AI007180 and F31 HL137304 (to M.D.K.); and pilot awards from the NYU CTSA grant UL1TR001445 from the National Center for Advancing Translational Sciences (NCATS) (to K.C.) and NYU Cancer Center grant P30CA016087 (to K.C.). Cell sorting/flow cytometry (NYU Langone Health’s Cytometry and Cell Sorting Laboratory), mass spectrometry (The Proteomics Labratory at NYU Langone Health) and electron-microscopy studies (The Microscopy Labratory at NYU Langone Health) are supported in part by NYU Langone Health’s Laura and Isaac Perlmutter Cancer Center Support (grant P30CA016087) from the National Cancer Institute Langone. This work was also supported by a Faculty Scholar grant from the Howard Hughes Medical Institute, the Merieux Institute, the Kenneth Rainin Foundation, the Crohn’s & Colitis Foundation and the Stony Wold-Herbert Fund (all to K.C.). K.C. and V.J.T. are Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases.
Author information
Authors and Affiliations
Contributions
M.D.K., V.J.T. and K.C. formulated the original hypothesis and designed the study. M.D.K. preformed the experiments and analysed the data. K.L.C. provided assistance with experiments that examined vesicle-trafficking events downstream of TLR9 activation. F.-X.L. provided assistance with TEM, and A.D. and B.M.U. performed and assisted in the analysis of mass-spectrometry experiments. K.T. generated the construct to express α-toxin, and purified and characterized the toxin. D.U. provided essential reagents and insight into the shRNA transduction experiments. M.D.K, V.J.T. and K.C. wrote the manuscript. All authors commented on the manuscript, data and conclusions.
Corresponding authors
Ethics declarations
Competing interests
V.J.T. is an inventor on patents and patent applications filed by NYU, which are currently under commercial license to Janssen Biotech. K.C. has consulted for, or received an honorarium from, Puretech Health, Genentech and AbbVie; has received research support from Puretech Health, Pacific Biosciences and Pfizer; and has a provisional patent, US Patent application number 15/625,934.
Additional information
Peer review information Nature thanks Ivan Dikic, Lora Hooper, Andreas Peschel, Philip Stahl and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 ADAM10 and EpCAM levels following lysosomal inhibition with ammonium chloride, chloroquine or bafilomycin or proteasomal inhibition with MG132.
a, Time course of flow-cytometry analysis of ADAM10 following lysosomal inhibition with ammonium chloride (NH4Cl, 20 mM), chloroquine (CQ, 50 μM) or PBS as a control; n = 3. b–d, Western blot analysis of ADAM10 and SQSTM1 following lysosomal inhibition with NH4Cl, CQ or bafilomycin (BAF, 10 nM). Shown are a representative western blot from four independent experiments (b), quantification of ADAM10 levels (n = 5) (c) and quantification of SQSTM1 levels (n = 3) (d) at 24 h after inhibition. e, f, Representative histogram (e) and quantification (f) of cell-surface EpCAM in BAF-treated A549 cells; n = 3. g, Time course of flow-cytometry analysis of EpCAM following treatment with NH4Cl or CQ; n = 4. h, i, ADAM10, P4D1 and actin levels following proteasomal inhibition with the chemical compound MG132. Shown are a flow-cytometry time course of cell-surface ADAM10 levels following MG132 treatment (h) and a representative western blot from three independent experiments (i); n = 3. Measurements were taken from distinct samples and graphs show mean ± s.e.m. a, c, d, f–h, One-way ANOVA with Dunnet’s post-test compared with PBS treatment or time 0.
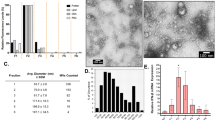
Extended Data Fig. 2 Exosome-isolation and quantification strategies.
a, Exosome-isolation protocol from in vitro or in vivo sources. Exosomes are isolated using a multistep centrifugation procedure including a 0.22-μm filtration step. b, Western blot of actin, ARF6 and CD9 following each sequential centrifugation step during exosome isolation. c, Electron-microscopy (EM) quantification of vesicles 80–150 nm and greater than 150 nm in size; n = 80 images. d, EM negative staining of exosome fractions. Arrows indicate exosomes and protein aggregates. e, Representative EM images of the exosome fraction, and zoomed insets with arrows indicating the single membranes of exosomes. f, Gating strategy and representative flow-cytometry plots from nt shRNA and ATG16L1 KD samples of six independently repeated experiments. Exosomes were stained with antibodies against CD9, CD63, CD81 and ADAM10. Exosomes were concurrently labelled with PKH67, a lipid-membrane-incorporating dye. FSC, forward scatter; SSC, side scatter. Measurements were taken from distinct samples and graphs show means ± s.e.m. c, Two-tailed, unpaired t-test with Welch’s correction compared with PBS controls.
Extended Data Fig. 3 CQ and BAF elicit production of ADAM10-positive exosomes.
a–d, Western blot analysis of cell lysate CD9 (cell CD9), exosome CD9 (exo CD9) and exosome ADAM10 (exo ADAM10) following addition of CQ or BAF. Shown are a representative western blot from six independent experiments (a) and quantification of cell CD9 (b), exosome CD9 (c), and exosome ADAM10 (d) after PBS, CQ or BAF treatment. Measurements were taken from distinct samples, and graphs show means ± s.e.m. b–d, One-way ANOVA with Dunnet’s post-test compared with PBS controls.
Extended Data Fig. 4 Exosomes are produced in response to bacterial exposure.
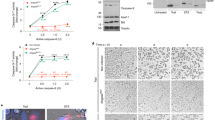
a, Flow-cytometry quantification of exosomes per 100,000 events in mouse BMDCs and BMDMs with or without exposure to HKSA (BMDCs plus PBS, n = 6; BMDCs plus HKSA, n = 6; BMDMs plus PBS, n = 3; BMDMs plus HKSA, n = 4). b–h, Quantification of total exosomes in A549 cell-culture supernatant by flow cytometry 18 h after treatment with peptidoglycan (PDG; b), lipopolysaccharide (LPS; c), lipoteichoic acid (LTA; d), Pam3CSK (e), Pam2CSK (f) or S. aureus RNA (SA RNA; g); n = 3. h, Quantification of total exosomes in TLR9 KD A549 cell-culture supernatants following treatment with CpG DNA; n = 3. i, Flow-cytometry quantification of A549-produced exosomes following exposure to HKSA or to a strain of S. aureus deficient in the production of α-toxin (HK dHLA). j, Flow-cytometry quantification of exosomes isolated from A549 cells treated with PBS (n = 3), S. aureus genomic DNA (SA gDNA; 0.5 μg ml−1; n = 5), and/or DNase I (n = 2). k, Flow-cytometry quantification of exosomes isolated from cells treated with BAF (n = 5), Torin-1 (n = 6), or both (n = 3). l, Representative western blot of SQSMT1, LC3I/II and actin in cells treated with BAF, Torin-1 or both 4 h after treatment, from two independent experiments. m, Flow-cytometry quantification of exosomes from A549 cells treated with PBS (mock treatment; n = 8), CpG DNA (n = 8), or CpG DNA and GW4869 (n = 7). n, o, Plasma exosome quantification of ATG16L1 flow/flox; CD11c-Cre (n), and ATG16L1 flow/flox; LysM-Cre (o) following exposure to either CpG DNA or HKSA, respectively. p, Representative western blot of ADAM10, ASS1, CD9 and CD81 in exosome fractions submitted to mass spectrometry, from three independent experiments. Measurements were taken from distinct samples and graphs show means ± s.e.m. a, b–h, Two-tailed, unpaired t-test with Welch’s correction compared with PBS controls. j, k, m–o, One-way ANOVA with Dunnet’s post-test compared with PBS, mock, CpG DNA or Cre−/+ controls.
Extended Data Fig. 5 BAF and CpG DNA decrease acidic organelles.
a–c, Representative flow-cytometry histograms from three independent experiments of Lysosensor signal following treatment with BAF or CpG DNA (a), or BAF with or without CpG DNA (b). c, Quantification of Lysosensor MFI following treatment with PBS alone (no BAF or CpG DNA; n = 8), BAF (n = 6), CpG DNA (n = 9), or BAF plus CpG DNA (n = 6) (c). Measurements were taken from distinct samples and graphs show means ± s.e.m. c, One-way ANOVA with Dunnet’s post-test compared with PBS controls.
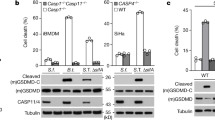
Extended Data Fig. 6 Exosomes protect from S. aureus toxicity in vitro and in vivo.
a, Flow-cytometry exosome quantification from nt shRNA control and ADAM10 KD A549 cells; n = 3. b, Cell death, measured by LDH release, of A549 cells treated with α-toxin only, pretreated with HKSA or CpG DNA and α-toxin (‘induced’), or pre-exposed to HKSA or CpG DNA followed by PBS wash and then α-toxin treatment (‘induced; washed’); n = 5. c, Representative flow-cytometry histograms of CCR5 on CD81-positive, CD63-positive and CD9-positive exosomes isolated from mouse BMDMs. d, Exogenous exosome-transfer protocol. In step 1, donor mice are pre-exposed to HKSA i.v. to induce exosome production. In step 2, exosomes from donor mice are injected intraperitoneally (i.p.) on day −1, day 0 and day +1 following lethal i.v. injection of S. aureus. e, Survival of wild-type mice infected i.v. with a lethal dose of 5 × 107 CFU of S. aureus (USA300) that were mock-treated (n = 10) or injected i.p. with exosomes from Atg16l1HM mice (n = 8). f, Endogenous exosome-protection protocol. Mice are i.v. injected with HKSA to induce exosome production. Four hours later, mice are infected with a lethal dose of S. aureus (2.5–5 × 107). g, h, Western blot analysis of α-toxin oligomerization in total BAL or in exosome fraction in BAL of mice pre-exposed to HKSA or PBS intranasally (i.n.), representative of four independent experiments. i, j, Quantification of α-toxin monomer (i) and heptamer (j) in BAL and exosome fraction following pre-exposure; n = 4. k, Ratio of α-toxin heptamer in exosome fraction to total α-toxin signal in BAL; n = 4. Measurements were taken from distinct samples and graphs show means ± s.e.m. a, i–k, Two-tailed, unpaired t-test with Welch’s correction. b, One-way ANOVA with Dunnet’s post-test compared with α-toxin only or ‘induced’ controls. e, log-rank Mantel–Cox test.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 and Supplementary Table 1-3.
Rights and permissions
About this article
Cite this article
Keller, M.D., Ching, K.L., Liang, FX. et al. Decoy exosomes provide protection against bacterial toxins. Nature 579, 260–264 (2020). https://doi.org/10.1038/s41586-020-2066-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-2066-6
This article is cited by
-
The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections
Inflammation and Regeneration (2024)
-
Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer
Experimental & Molecular Medicine (2024)
-
Soluble receptors in cancer: mechanisms, clinical significance, and therapeutic strategies
Experimental & Molecular Medicine (2024)
-
Secretion of the fungal toxin candidalysin is dependent on conserved precursor peptide sequences
Nature Microbiology (2024)
-
Host extracellular vesicles confer cytosolic access to systemic LPS licensing non-canonical inflammasome sensing and pyroptosis
Nature Cell Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.