Abstract
The mammalian claustrum, owing to its widespread connectivity with other forebrain structures, has been hypothesized to mediate functions that range from decision-making to consciousness1. Here we report that a homologue of the claustrum, identified by single-cell transcriptomics and viral tracing of connectivity, also exists in a reptile—the Australian bearded dragon Pogona vitticeps. In Pogona, the claustrum underlies the generation of sharp waves during slow-wave sleep. The sharp waves, together with superimposed high-frequency ripples2, propagate to the entire neighbouring pallial dorsal ventricular ridge (DVR). Unilateral or bilateral lesions of the claustrum suppress the production of sharp-wave ripples during slow-wave sleep in a unilateral or bilateral manner, respectively, but do not affect the regular and rapidly alternating sleep rhythm that is characteristic of sleep in this species3. The claustrum is thus not involved in the generation of the sleep rhythm itself. Tract tracing revealed that the reptilian claustrum projects widely to a variety of forebrain areas, including the cortex, and that it receives converging inputs from, among others, areas of the mid- and hindbrain that are known to be involved in wake–sleep control in mammals4,5,6. Periodically modulating the concentration of serotonin in the claustrum, for example, caused a matching modulation of sharp-wave production there and in the neighbouring DVR. Using transcriptomic approaches, we also identified a claustrum in the turtle Trachemys scripta, a distant reptilian relative of lizards. The claustrum is therefore an ancient structure that was probably already present in the brain of the common vertebrate ancestor of reptiles and mammals. It may have an important role in the control of brain states owing to the ascending input it receives from the mid- and hindbrain, its widespread projections to the forebrain and its role in sharp-wave generation during slow-wave sleep.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequencing data have been deposited in the NCBI Sequence Read Archive: BioProjects PRJNA591493 (lizard) and PRJNA408230 (turtle). Links to those archives and to analysis code can be found at: https://brain.mpg.de/research/laurent-department/software-techniques.html. Data are also available from the corresponding author on request.
References
Crick, F. C. & Koch, C. What is the function of the claustrum? Phil. Trans. R. Soc. Lond. B 360, 1271–1279 (2005).
Buzsáki, G. Hippocampal sharp wave-ripple: a cognitive biomarker for episodic memory and planning. Hippocampus 25, 1073–1188 (2015).
Shein-Idelson, M., Ondracek, J. M., Liaw, H. P., Reiter, S. & Laurent, G. Slow waves, sharp waves, ripples, and REM in sleeping dragons. Science 352, 590–595 (2016).
Saper, C. B. & Fuller, P. M. Wake-sleep circuitry: an overview. Curr. Opin. Neurobiol. 44, 186–192 (2017).
Weber, F. & Dan, Y. Circuit-based interrogation of sleep control. Nature 538, 51–59 (2016).
Scammell, T. E., Arrigoni, E. & Lipton, J. O. Neural circuitry of wakefulness and sleep. Neuron 93, 747–765 (2017).
Lyamin, O. I., Manger, P. R., Ridgway, S. H., Mukhametov, L. M. & Siegel, J. M. Cetacean sleep: an unusual form of mammalian sleep. Neurosci. Biobehav. Rev. 32, 1451–1484 (2008).
Naumann, R. K. & Laurent, G. in Evolution of Nervous Systems Vol. 1 (ed. Kaas, J. H.) 491–518 (Elsevier, 2017).
Moreno, N. & González, A. Evolution of the amygdaloid complex in vertebrates, with special reference to the anamnio-amniotic transition. J. Anat. 211, 151–163 (2007).
Puelles, L. et al. in Evolution of Nervous Systems Vol. 1 (ed. Kaas, J. H.) 519–555 (Elsevier, 2017).
Tosches, M. A. et al. Evolution of pallium, hippocampus, and cortical cell types revealed by single-cell transcriptomics in reptiles. Science 360, 881–888 (2018).
Wang, Q. et al. Organization of the connections between claustrum and cortex in the mouse. J. Comp. Neurol. 525, 1317–1346 (2017).
Saunders, A. et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell 174, 1015–1030 (2018).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Striedter, G. F. The telencephalon of tetrapods in evolution. Brain Behav. Evol. 49, 179–213 (1997).
Monti, J. M. Serotonin control of sleep-wake behavior. Sleep Med. Rev. 15, 269–281 (2011).
Clément, O., Sapin, E., Bérod, A., Fort, P. & Luppi, P. H. Evidence that neurons of the sublaterodorsal tegmental nucleus triggering paradoxical (REM) sleep are glutamatergic. Sleep 34, 419–423 (2011).
Hobson, J. A., McCarley, R. W. & Wyzinski, P. W. Sleep cycle oscillation: reciprocal discharge by two brainstem neuronal groups. Science 189, 55–58 (1975).
da Costa, N. M., Fürsinger, D. & Martin, K. A. The synaptic organization of the claustral projection to the cat’s visual cortex. J. Neurosci. 30, 13166–13170 (2010).
Druga, R. in The Claustrum: Structural, Functional, and Clinical Neuroscience (eds Smythies, J. R. et al.) 29–84 (Academic, 2014).
Olson, C. R. & Graybiel, A. M. Sensory maps in the claustrum of the cat. Nature 288, 479–481 (1980).
Pammer, L. Explorations of Turtle Cortex Function through Molecular, Optogenetic and Electrophysiological Techniques. PhD thesis, Goethe Univ. (2017).
Tervo, D. G. et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron 92, 372–382 (2016).
Oh, S. W. et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014).
Harris, J. A., Oh, S. W. & Zeng, H. Adeno-associated viral vectors for anterograde axonal tracing with fluorescent proteins in nontransgenic and Cre driver mice. Curr. Protoc. Neurosci. 59, 1.20.1–1.20.18 (2012).
Desan, P. H. in The Forebrain of Reptiles (eds Schwerdtfeger, W. K. & Smeets, W. J.) 1–11 (Karger, 1987).
Heller, S. B. & Ulinski, P. S. Morphology of geniculocortical axons in turtles of the genera Pseudemys and Chrysemys. Anat. Embryol. 175, 505–515 (1987).
Atlan, G. et al. The claustrum supports resilience to distraction. Curr. Biol. 28, 2752–2762 (2018).
Smythies, J., Edelstein, L. & Ramachandran, V. Hypotheses relating to the function of the claustrum. Front. Integr. Neurosci. 6, 53 (2012).
Dillingham, C. M., Janowski, M. M., Chandra, R., Frost, B. E. & O’Mara, S. M. The claustrum: considerations regarding its anatomy, functions and a programme for research. Brain Neurosci. Adv. 1, 1–9 (2017).
Edelstein, L. R. & Denaro, F. J. The claustrum: a historical review of its anatomy, physiology, cytochemistry and functional significance. Cell. Mol. Biol. 50, 675–702 (2004).
Goll, Y., Atlan, G. & Citri, A. Attention: the claustrum. Trends Neurosci. 38, 486–495 (2015).
Mathur, B. N., Caprioli, R. M. & Deutch, A. Y. Proteomic analysis illuminates a novel structural definition of the claustrum and insula. Cereb. Cortex 19, 2372–2379 (2009).
Puelles, L. in The Claustrum: Structural, Functional, and Clinical Neuroscience (eds Smythies, J. R. et al.) 119–176 (Academic, 2014).
Briscoe, S. D., Albertin, C. B., Rowell, J. J. & Ragsdale, C. W. Neocortical association cell types in the forebrain of birds and alligators. Curr. Biol. 28, 686–696 (2018).
Buchanan, K. J. & Johnson, J. I. Diversity of spatial relationships of the claustrum and insula in branches of the mammalian radiation. Ann. NY Acad. Sci. 1225, E30–E63 (2011).
Gabor, A. J. & Peele, T. L. Alterations of behavior following stimulation of the claustrum of the cat. Electroencephalogr. Clin. Neurophysiol. 17, 513–519 (1964).
Renouard, L. et al. The supramammillary nucleus and the claustrum activate the cortex during REM sleep. Sci. Adv. 1, e1400177 (2015).
Jackson, J., Karnani, M. M., Zemelman, B. V., Burdakov, D. & Lee, A. K. Inhibitory control of prefrontal cortex by the claustrum. Neuron 99, 1029–1039 (2018).
Narikiyo, K. et al. The claustrum coordinates cortical slow-wave activity. Preprint at bioRxiv https://doi.org/10.1101/286773 (2018).
Siapas, A. G. & Wilson, M. A. Coordinated interactions between hippocampal ripples and cortical spindles during slow-wave sleep. Neuron 21, 1123–1128 (1998).
McInnes, L., Healy, J. & Melville, J. UMAP: Uniform manifold approximation and projection for dimension reduction. Preprint at http://arxiv.org/abs/1802.03426 (2018).
Moreno, N., Domínguez, L., Morona, R. & González, A. Subdivisions of the turtle Pseudemys scripta hypothalamus based on the expression of regulatory genes and neuronal markers. J. Comp. Neurol. 520, 453–478 (2012).
Medina, L., Smeets, W. J., Hoogland, P. V. & Puelles, L. Distribution of choline acetyltransferase immunoreactivity in the brain of the lizard Gallotia galloti. J. Comp. Neurol. 331, 261–285 (1993).
Bruce, L. L. & Neary, T. J. Afferent projections to the ventromedial hypothalamic nucleus in a lizard, Gekko gecko. Brain Behav. Evol. 46, 14–29 (1995).
Bruce, L. L. & Neary, T. J. Afferent projections to the lateral and dorsomedial hypothalamus in a lizard, Gekko gecko. Brain Behav. Evol. 46, 30–42 (1995).
Ebner, F. F. in Evolution of Brain and Behavior in Vertebrates (eds Masterton, R. B. et al.) 115–167 (Taylor & Francis, 1976).
Font, C., Lanuza, E., Martinez-Marcos, A., Hoogland, P. V. & Martinez-Garcia, F. Septal complex of the telencephalon of lizards: III. Efferent connections and general discussion. J. Comp. Neurol. 401, 525–548 (1998).
Hoogland, P. V. & Vermeulen-Vanderzee, E. Efferent connections of the dorsal cortex of the lizard Gekko gecko studied with Phaseolus vulgaris–leucoagglutinin. J. Comp. Neurol. 285, 289–303 (1989).
Smeets, W. J. & Steinbusch, H. W. Distribution of noradrenaline immunoreactivity in the forebrain and midbrain of the lizard Gekko gecko. J. Comp. Neurol. 285, 453–466 (1989).
Smeets, W. J., Hoogland, P. V. & Voorn, P. The distribution of dopamine immunoreactivity in the forebrain and midbrain of the lizard Gekko gecko: an immunohistochemical study with antibodies against dopamine. J. Comp. Neurol. 253, 46–60 (1986).
ten Donkelaar, H. J., Bangma, G. C., Barbas-Henry, H. A., de Boer-van Huizen, R. & Wolters, J. G. The brain stem in a lizard, Varanus exanthematicus. Adv. Anat. Embryol. Cell Biol. 107, 1–2 (1987).
ten Donkelaar, H. J. in The Central Nervous System of Vertebrates Vol. 1–3 (eds Nieuwenhuys, H. et al.) 1315–1524 (Springer, 1998).
Wolters, J. G., ten Donkelaar, H. J., Steinbusch, H. W. & Verhofstad, A. A. Distribution of serotonin in the brain stem and spinal cord of the lizard Varanus exanthematicus: an immunohistochemical study. Neuroscience 14, 169–193 (1985).
Wolters, J. G., ten Donkelaar, H. J. & Verhofstad, A. A. Distribution of catecholamines in the brain stem and spinal cord of the lizard Varanus exanthematicus: an immunohistochemical study based on the use of antibodies to tyrosine hydroxylase. Neuroscience 13, 469–493 (1984).
Pedersen, N. P. et al. Supramammillary glutamate neurons are a key node of the arousal system. Nat. Commun. 8, 1405 (2017).
Acknowledgements
We thank K. Steele and A. Schwartzlose for help with serotonin immunocytochemistry and neural tract tracing; E. Northrup and G. Wexel for veterinary care; T. Klappich for reptile care; S. Junek, F. Vollrath and C. Polisseni for help with imaging and microscopy; G. Tushev for help with genome reannotation; E. Desfilis for help with lizard neuroanatomy; and members of the Laurent laboratory for help and comments. The work was funded by the Max Planck Society, the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) (ERC grant agreement no. 322705), the European Research Council under the European Union’s Horizon 2020 research and innovation programme (ERC grant agreement no. 834446) and the DFG (CRC1080) (G.L.); postdoctoral fellowships from the JSPS (SPD and for research abroad) and from the Kanae Foundation for the promotion of medical science (H.N.); and an EMBO long-term fellowship (ALTF 421-2017) (L.A.F.).
Author information
Authors and Affiliations
Contributions
H.N. and L.A.F. contributed equally and have equal right to list themselves first in bibliographic documents. H.-H.L. and M.A.T. also contributed equally. Project conception, H.N., L.A.F. and G.L.; animal surgery, M.K., H.N. and L.A.F.; electrophysiology, H.N., L.A.F. and S.R.; pharmacology, H.N., R.K. and L.A.F.; scRNA-seq, T.G.F., D.H., A.M. and M.A.T.; bioinformatics, M.A.T. and D.H.; tracing, anatomy and histology, L.A.F., H.-H.L., R.K., T.G.F., A.A. and M.K.; experimental design, data interpretation and analysis, H.N., L.A.F., H.-H.L., M.A.T., T.G.F., D.H., S.R. and G.L.; project management and supervision, G.L.; manuscript writing, G.L., with input from all co-authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Michael Lazarus and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Further description of SWR statistics and propagation in vivo.
a, The amplitude and frequency of sharp waves vary as the animal transitions between slow-wave and REM sleep. Top, illustrative LFP trace (<150 Hz) showing a decrease in sharp-wave amplitude and frequency around the slow-wave–REM transition point. Open circles indicate detected sharp waves3 (see Methods). Data in a–d are from the same animal and a single night, and correspond to the recording in Fig. 1 (anterior recording site, red). Statistics are based on n = 11,123 sharp waves. b, Distribution of sharp-wave width (measured at half peak amplitude) and peak amplitude from the animal in a and Fig. 1. Pr, probability. c, Average sharp-wave trace ± 1 s.d. (grey) calculated over n = 11,123 sharp waves. d, Inter-event interval (IEI) for sharp waves recorded during slow-wave sleep. The y axis (probability) is on a logarithmic scale. e, f, Summary of data recorded over five nights from two animals. Each circle represents the mean of one night; black line shows the median. e, Mean inter-event intervals during slow-wave sleep. f, Mean sharp-wave width and amplitude (n = 8,055–13,494 sharp waves per night). g, Delay distributions of sharp waves in anterior (or posterior) DVR, triggered on simultaneously recorded posterior (or anterior) DVR. Sharp waves from three nights (animal 1; n = 24,501 sharp waves) and two nights (animal 2; n = 13,070 sharp waves). h, Locations of simultaneous recording sites in the aDVR (circles). Left, schematic of recording configuration. Middle and right, confocal images highlighting the recording sites, as identified by electrolytic lesions and DiI dye that was applied to the back of the silicon probes. Post hoc staining with an antibody against hippocalcin was used to determine the borders of the claustrum (see Fig. 3).
Extended Data Fig. 2 Comparison of SWR statistics across preparations and recording conditions.
a, Slice preparation (see Methods) for field-potential recordings. b, Spontaneous sharp waves (LFP; <150 Hz) and corresponding ripples (high-pass (HP) band; 70–150 Hz) in the amDVR. Insets: top left, magnification of the SWR marked with a dotted box; top right, 350 ripples; high-pass signal intensity (HPI) >70 Hz aligned on trough of sharp wave (overlaid as average). c, Distribution of amplitude (x) and width (y, full width at half maximum) of SWR events in a representative DVR slice. d, Distribution of SWR amplitude and width (as in c) in a representative ex vivo preparation. e, Ratio of amplitude (μV) to width (ms); n = 5 sleep epochs from 3 animals (in vivo; blue), 4 ex vivo brains (red) and 12 slices (green). Lines show the mean. f, Autocorrelation function of sharp-wave times, showing that the characteristic rhythmic modulation of sharp-wave generation (which is due to the alternation of slow-wave sleep and REM sleep with a 2–3 min period) in sleeping animals is absent from both ex vivo brain preparations and slice preparations (n = 5 sleep epochs from 3 animals (in vivo), 4 ex vivo brains and 12 slices). g, Whole-cell patch-clamp recording (in current-clamp mode) of a DVR neuron (Vm), together with LFP recording in a neighbouring region (V (LFP)) with a glass micropipette. Note the simultaneous depolarization of the neuron and SWRs, and moderate neuronal depolarization that gives rise to occasional firing (three action potentials here). The experiment was repeated with 12 neurons. h, Whole-cell patch-clamp recording of an amDVR neuron in voltage-clamp mode, held at depolarized (blue) and hyperpolarized (red) holding potentials (Vh). Note the volleys of excitatory (red) and inhibitory (blue) currents at each SWR (LFP), and the near absence of synaptic input in between. i, Spike times of a patched amDVR neuron in relation to sharp waves. Note the locking to the sharp-wave trough (t = 0), and the absence of firing otherwise (n = 2 amDVR neurons). j, Mean excitatory (ge) and inhibitory (gi) conductances (n = 20 and 21 events, respectively). The black and grey lines show averaged sharp waves recorded with inhibitory and excitatory conductances, respectively. Traces are aligned on the sharp-wave trough.
Extended Data Fig. 3 Additional single-cell transcriptomic characterization.
a, UMAP42 representation of 20,257 Pogona telencephalic cells, colour-coded by cluster. EG, ependymoglial cells; ExcNeur, excitatory neurons; InhNeur, inhibitory neurons; MG, microglia; mur, mural cells; NPC, neural progenitor cells; olig, oligodendrocytes; OPC, oligodendrocyte progenitor cells; prol, proliferating cells; RBC, red blood cells. b, Dot plot showing the expression of canonical cell markers (rows) across telencephalic cell clusters (columns). The size of the dot corresponds to the percentage of cells in a cluster in which the gene has been detected, and the colour represents the expression level. c, UMAP representation of 9,777 lizard telencephalic neurons, colour-coded by cluster. d, e, UMAP representations of glutamatergic (slc17a7) and GABAergic (slc32a1) neurons in the telencephalon dataset. f, Double colorimetric in situ hybridization in a frontal section through the anterior Pogona forebrain. Scale bar, 1 mm. slc32a1 (blue) labels GABAergic neurons in the subpallium and scattered GABAergic neurons that have migrated from subpallium to pallium. slc17a6 (orange) labels glutamatergic neurons in the pallial region. g, Ordered matrix of pairwise Pearson correlations between the expression of 143 ion-channel and neurotransmitter-receptor genes detected in this glutamatergic pallial dataset from Pogona (see Extended Data Fig. 5). The dendrogram (top) is based on correlation coefficients and Ward.D2 linkage; red indicates a gene module with enriched expression in the amDVR. h, Average expression, in the 29 glutamatergic Pogona clusters, of the 143 genes in g (and Extended Data Fig. 5). Genes with enriched expression in the amDVR are listed on the right, with relevant neurotransmitter receptor genes in bold. i, UMAP representation of 4,054 lizard pallial glutamatergic neurons, colour-coded by cluster (same as in Fig. 3a). j, Dot plot showing the expression of specific cluster markers (rows) in the 29 pallial glutamatergic clusters (columns). The size of the dot corresponds to the percentage of cells in a cluster in which the gene has been detected, and the colour represents the expression level.
Extended Data Fig. 4 Mini-slices of the DVR and localization of SWR generation.
a, Left, recording configuration of mini-slices of the DVR on a planar 252-channel microelectrode array. Dots represent electrodes. Right, post hoc immunostaining of the mini-slices. Red, Nissl; green, hippocalcin. b, Left, spatial distribution of SWR waveforms as recorded from the mini-slices in a. Right, illustrative LFP traces recorded from the amDVR or claustrum (1) and plDVR (2) (see recording positions on the microelectrode array on the left). In conclusion, SWRs occur spontaneously in the amDVR, and are absent from the plDVR once it is disconnected from the amDVR (claustrum).
Extended Data Fig. 5 Ion-channel and neurotransmitter-receptor mRNAs in the glutamatergic cell clusters of the Pogona telencephalon.
a, Dot plot showing expression of ion-channel and neurotransmitter-receptor genes (rows) in Pogona glutamatergic clusters (columns 1–29). The plot shows only genes that were detected in at least 20% of the cells of at least one cluster. The size of the dot corresponds to the percentage of cells in a cluster in which the gene has been detected, and the colour represents the expression level. Clusters 19 and 20 (box) correspond to the amDVR or claustrum. They differ by the expression of some acetylcholine- and serotonin-receptor subtypes (see also Fig. 3h). b, Ordered pairwise Pearson correlation matrix of cluster transcriptomes, calculated from the expression of the ion-channel and neurotransmitter-receptor genes in a. This gene set is sufficient to distinguish the amDVR clusters (19 and 20) from all of the others. The dendrogram is based on Pearson correlations and Ward.D2 linkage.
Extended Data Fig. 6 Identification of potential regulatory areas of brain states and distribution of GFP-labelled neurons after injection of rAAV2-retro into the claustrum.
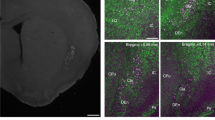
a, Left, schematic of the Pogona brain in sagittal view, showing the regions defined by immunohistochemistry, in situ hybridization and retrograde tracing. Numbers 1–7 indicate the levels of the transverse sections that are shown on the right. Right (panels 1–7), micrographs and corresponding schematic representations of relevant areas (in red), identified by immunohistochemistry, in situ hybridization and Nissl staining. Scale bars, 500 μm. Far right of panels 1–7, magnified views of area(s) delineated as box(es) in the corresponding photomicrographs. Scale bars, 100 μm. b, Identification of rAAV2-retro injection sites. Scale bars, 500 μm. The red channel is not shown in the rightmost image. c, Illustrative examples of retrograde labelling of claustrum connectivity, in transverse sections. Panels 1, 2, inputs to claustrum revealed by rAAV2-retro injection in the claustrum. Panel 1, injection site in lateral claustrum. The claustrum is indicated by anti-hippocalcin immunostain (pink). Note retro-labelled cells in the anterior dorsal cortex (box, magnified at right). Panel 2, same brain as in 1, but a more posterior section. The labelled region in the box is the dorsal lateral amygdala. Panels 3–12, representative images illustrating the distribution of GFP-labelled neurons in the DLPT, DLT, DMT, prethalamus, SUM, mammillary nucleus (MN), TMN, VTA, SN, PAG, LoC and SC, with projections to the claustrum. Abbreviations as in Fig. 3. The catecholaminergic neuron marker tyrosine hydroxylase (TH) was used to indicate the location of the VTA, SN and LoC. Scale bars, 500 μm. Scale bars for magnified areas: DLPT, DLT, DMT, prethalamus, SUM, MN, TMN, VTA, LoC, 50 μm; SN, PAG, SC, 100 μm.
Extended Data Fig. 7 The claustrum of lizard and turtle differ in position and architectonics, but are both autonomous sources of SWRs.
a, Transcriptomic similarity between turtle and lizard clusters, measured as the fraction of single cells that mapped from the turtle pallium dataset to the Pogona clusters (Methods). Note that the turtle cell clusters e03–e06 (pallial thickening; PT) map to the lizard cluster 19 (amDVR or claustrum). Turtle data and clusters are from a previous study11. b, In situ hybridization in an anterior transverse section, showing expression of the pallial thickening marker gene crhbp. Scale bar, 500 μm. c, Architectonics of the lizard claustrum. Right, retrograde labelling of claustrum neurons by rAAV2-retro injected into the aDVR. Left, magnification of the boxed area on the right (in the claustrum). Note the disordered distribution of multipolar neurons. Pink colour shows anti-hippocalcin immunostaining. Scale bars, 100 μm (left); 500 μm (right). d, Architectonics of the turtle claustrum. Right, retrograde labelling of claustrum neurons by rAAV2-retro injected into the dorso-medial cortex. Left, magnification of the boxed area on the right. Note the arrangement of bipolar neurons within the pallial thickening layer (see also b for layering of pallial thickening). Scale bars, 100 μm (left); 500 μm (right). e, Spontaneous sharp waves recorded simultaneously in the claustrum and the DVR in turtle slice preparation. The red dots in the schematic indicate recording sites. Note sharp wave (LFP) and ripple in the high-pass (HP) band. f, Bottom, 295 successive spontaneous ripples (high-pass signal intensity (HPI) > 70 Hz) aligned on the trough of each sharp wave. Top, average of 295 sharp waves aligned on waveform troughs. Grey shading represents s.d. g, Representative cross-correlogram of LFP traces recorded simultaneously from the claustrum and the DVR (with claustrum as reference), showing the sharp waves from the DVR trailing those from the claustrum.
Extended Data Fig. 8 SWR recordings and stimulation experiments with lizard ex vivo brain preparations.
a–f, Experiments in ex vivo brain preparation after cortex removal. a, Top, ex vivo brain preparation. Bottom, spontaneous SWRs recorded in the claustrum (<150 Hz) (top trace); HP: 70–150-Hz filtered LFP, showing ripples (bottom trace). b, Local pressure injection of 20 μM TTX into the claustrum and post hoc assessment of injection with Evans blue (transverse section at the bottom, red). c, Injection of TTX into the claustrum (shading) silences sharp-wave activity in the claustrum, but also (indirectly) in the DVR. d, Analysis of four experiments as in c. The filled circles represent mean ± s.e.m. Claustrum: *P = 0.029, T = 26 (two-sided Mann–Whitney rank-sum test); DVR: *P = 0.029, T = 26 (two-sided Mann–Whitney rank-sum test). e, Top, average trace and s.d. (shading) from 3,842 sharp waves recorded from the claustrum of an ex vivo forebrain (alignment on trough). Bottom, HPI (>70 Hz) aligned on sharp-wave trough, showing ripple alignment. f, Top, simultaneous recordings from ipsilateral claustrum and DVR in an ex vivo preparation. Bottom, cross-correlation between simultaneous recordings in ipsilateral claustrum and DVR, showing that the claustrum precedes the DVR by around 100 ms. g, Peristimulus time histogram for multi-unit activity in the cortex, in response to activation of ipsilateral claustrum in an intact ex vivo forebrain. The experiment was carried out in normal ACSF at room temperature in the presence of 30 μM serotonin to suppress spontaneous SWRs in the claustrum and 50 μM carbachol to raise cortex excitability. The claustrum stimulus consisted of a single 50-μs electrical pulse, delivered with a bipolar electrode. Cortex multi-unit activity was recorded with a glass micropipette. h, Change in cortical firing rate (FR) measured in a 200-ms bin after the claustrum stimulus versus a 200-ms bin before the stimulus (as in g, on each side of t = 0). The control column plots the firing-rate ratio measured in the experiment in g, and the GBZ + CGP column plots the results of the same experiment after addition of the GABA receptor antagonists gabazine (GBZ; 5 μM) and CGP52432 (CGP; 2 μM); n = 4 ex vivo brains from 3 animals each. The control experiment shows that stimulation of the claustrum has an immediate and reliable inhibitory effect on the cortex (#: significantly different from baseline, P = 0.017, t3 = 4.8 (two-sided paired t-test)). The stimulation experiment in GABA receptor antagonists shows that stimulation of the claustrum now slightly excites the cortex (**: significantly different from control, P = 2.0 × 10−3, t6 = −5.22 (two-sided Student’s t-test)), suggesting that projections from the claustrum both activate and inhibit cortical neurons, probably via direct excitatory projections and indirect inhibitory ones through interneurons (see rodent experiments in a previous study39). Short horizontal lines indicate mean.
Extended Data Fig. 9 Further analysis of in vivo ibotenic-acid-induced lesion experiments in sleeping Pogona.
a, Autocorrelation (top) and cross-correlation (bottom) of β-band activity in the left and right DVR during sleep in an animal with bilateral claustrum lesions (lesions are shown in d). Note that a periodic sleep rhythm (period of around 3 min here) remains after claustrum lesions and therefore does not seem to depend on claustrum integrity. b, c, Same as a, but with unilateral ibotenic-acid-induced lesion in two animals (I and II). The non-lesioned (sham) side was injected with the same volume of PBS vehicle but without ibotenic acid. Dotted line, sham; solid line, lesion. d, Nissl stains (1–3) of transverse sections of the brain of animals with bilateral lesions (shown also in Fig. 4b), at levels indicated in the schematic on the left. Note the claustral lesions (arrows in 1), which are visible as cell-body loss, and the recording sites in left (2) and right (3) DVRs (dotted circles).
Extended Data Fig. 10 Further data on serotonergic projections to claustrum and their effects on the generation of sharp-wave ripples.
a, Transverse section of claustrum double-labelled with DAPI (blue, nuclei) and serotonin (axonal fibres) antibodies. Note the dense meshwork of serotonergic fibres. Scale bar, 50 μm. b, Frequency of spontaneous SWRs in claustrum mini-slices as a function of superfused serotonin concentration. Red circles represent individual experiments (slices). Black points and lines are mean ± s.e.m.
Supplementary information
Rights and permissions
About this article
Cite this article
Norimoto, H., Fenk, L.A., Li, HH. et al. A claustrum in reptiles and its role in slow-wave sleep. Nature 578, 413–418 (2020). https://doi.org/10.1038/s41586-020-1993-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-1993-6
This article is cited by
-
A simple and reliable method for claustrum localization across age in mice
Molecular Brain (2024)
-
Reconstruction of macroglia and adult neurogenesis evolution through cross-species single-cell transcriptomic analyses
Nature Communications (2024)
-
Involvement of the claustrum in the cortico-basal ganglia circuitry: connectional study in the non-human primate
Brain Structure and Function (2024)
-
Comparative MRI analysis of the forebrain of three sauropsida models
Brain Structure and Function (2024)
-
Interhemispheric competition during sleep
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.