Abstract
Active matter consists of units that generate mechanical work by consuming energy1. Examples include living systems (such as assemblies of bacteria2,3,4,5 and biological tissues6,7), biopolymers driven by molecular motors8,9,10,11 and suspensions of synthetic self-propelled particles12,13,14. A central goal is to understand and control the self-organization of active assemblies in space and time. Most active systems exhibit either spatial order mediated by interactions that coordinate the spatial structure and the motion of active agents12,14,15 or the temporal synchronization of individual oscillatory dynamics2. The simultaneous control of spatial and temporal organization is more challenging and generally requires complex interactions, such as reaction–diffusion hierarchies16 or genetically engineered cellular circuits2. Here we report a simple technique to simultaneously control the spatial and temporal self-organization of bacterial active matter. We confine dense active suspensions of Escherichia coli cells and manipulate a single macroscopic parameter—namely, the viscoelasticity of the suspending fluid— through the addition of purified genomic DNA. This reveals self-driven spatial and temporal organization in the form of a millimetre-scale rotating vortex with periodically oscillating global chirality of tunable frequency, reminiscent of a torsional pendulum. By combining experiments with an active-matter model, we explain this behaviour in terms of the interplay between active forcing and viscoelastic stress relaxation. Our findings provide insight into the influence of bacterial motile behaviour in complex fluids, which may be of interest in health- and ecology-related research, and demonstrate experimentally that rheological properties can be harnessed to control active-matter flows17,18. We envisage that our millimetre-scale, tunable, self-oscillating bacterial vortex may be coupled to actuation systems to act a ‘clock generator’ capable of providing timing signals for rhythmic locomotion of soft robots and for programmed microfluidic pumping19, for example, by triggering the action of a shift register in soft-robotic logic devices20.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data supporting the findings of this study are included within the paper and its Supplementary Materials.
Code availability
The custom codes used in this study are available from the corresponding author upon request.
References
Marchetti, M. C. et al. Hydrodynamics of soft active matter. Rev. Mod. Phys. 85, 1143–1189 (2013).
Danino, T., Mondragon-Palomino, O., Tsimring, L. & Hasty, J. A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010).
Sokolov, A. & Aranson, I. S. Physical properties of collective motion in suspensions of bacteria. Phys. Rev. Lett. 109, 248109 (2012).
Wensink, H. H. et al. Meso-scale turbulence in living fluids. Proc. Natl Acad. Sci. USA 109, 14308–14313 (2012).
Chen, C., Liu, S., Shi, X. Q., Chaté, H. & Wu, Y. Weak synchronization and large-scale collective oscillation in dense bacterial suspensions. Nature 542, 210–214 (2017).
Saw, T. B. et al. Topological defects in epithelia govern cell death and extrusion. Nature 544, 212–216 (2017).
Kawaguchi, K., Kageyama, R. & Sano, M. Topological defects control collective dynamics in neural progenitor cell cultures. Nature 545, 327–331 (2017).
Keber, F. C. et al. Topology and dynamics of active nematic vesicles. Science 345, 1135–1139 (2014).
Wu, K.-T. et al. Transition from turbulent to coherent flows in confined three-dimensional active fluids. Science 355, eaal1979 (2017).
Huber, L., Suzuki, R., Krüger, T., Frey, E. & Bausch, A. R. Emergence of coexisting ordered states in active matter systems. Science 361, 255–258 (2018).
Prost, J., Jülicher, F. & Joanny, J. F. Active gel physics. Nat. Phys. 11, 111–117 (2015).
Palacci, J., Sacanna, S., Steinberg, A. P., Pine, D. J. & Chaikin, P. M. Living crystals of light-activated colloidal surfers. Science 339, 936–940 (2013).
Bricard, A., Caussin, J.-B., Desreumaux, N., Dauchot, O. & Bartolo, D. Emergence of macroscopic directed motion in populations of motile colloids. Nature 503, 95–98 (2013).
Yan, J. et al. Reconfiguring active particles by electrostatic imbalance. Nat. Mater. 15, 1095–1099 (2016).
Karig, D. et al. Stochastic Turing patterns in a synthetic bacterial population. Proc. Natl Acad. Sci. USA 115, 6572–6577 (2018).
Vicker, M. G. Eukaryotic cell locomotion depends on the propagation of self-organized reaction–diffusion waves and oscillations of actin filament assembly. Exp. Cell Res. 275, 54–66 (2002).
Giomi, L., Mahadevan, L., Chakraborty, B. & Hagan, M. F. Banding, excitability and chaos in active nematic suspensions. Nonlinearity 25, 2245 (2012).
Hemingway, E. J. et al. Active viscoelastic matter: from bacterial drag reduction to turbulent solids. Phys. Rev. Lett. 114, 098302 (2015).
Wehner, M. et al. An integrated design and fabrication strategy for entirely soft, autonomous robots. Nature 536, 451–455 (2016).
Preston, D. J. et al. Digital logic for soft devices. Proc. Natl Acad. Sci. USA 116, 7750–7759 (2019).
Wioland, H., Woodhouse, F. G., Dunkel, J., Kessler, J. O. & Goldstein, R. E. Confinement stabilizes a bacterial suspension into a spiral vortex. Phys. Rev. Lett. 110, 268102 (2013).
López, H. M., Gachelin, J., Douarche, C., Auradou, H. & Clément, E. Turning bacteria suspensions into superfluids. Phys. Rev. Lett. 115, 028301 (2015).
Bozorgi, Y. & Underhill, P. T. Effects of elasticity on the nonlinear collective dynamics of self-propelled particles. J. Non-Newton. Fluid Mech. 214, 69–77 (2014).
Li, G. & Ardekani, A. M. Collective motion of microorganisms in a viscoelastic fluid. Phys. Rev. Lett. 117, 118001 (2016).
Liu, Y., Jun, Y. & Steinberg, V. Concentration dependence of the longest relaxation times of dilute and semi-dilute polymer solutions. J. Rheol. 53, 1069–1085 (2009).
Ginoux, J. M. & Letellier, C. Van der Pol and the history of relaxation oscillations: toward the emergence of a concept. Chaos 22, 023120 (2012).
Sokolov, A., Aranson, I. S., Kessler, J. O. & Goldstein, R. E. Concentration dependence of the collective dynamics of swimming bacteria. Phys. Rev. Lett. 98, 158102 (2007).
Hemingway, E. J., Cates, M. E. & Fielding, S. M. Viscoelastic and elastomeric active matter: linear instability and nonlinear dynamics. Phys. Rev. E 93, 032702 (2016).
Warner, M. & Terentjev, E. M. Liquid Crystal Elastomers (Oxford Univ. Press, 2007).
Doostmohammadi, A., Ignés-Mullol, J., Yeomans, J. M. & Sagués, F. Active nematics. Nat. Commun. 9, 3246 (2018).
Aditi Simha, R. & Ramaswamy, S. Hydrodynamic fluctuations and instabilities in ordered suspensions of self-propelled particles. Phys. Rev. Lett. 89, 058101 (2002).
Murray, J. D. Mathematical Biology: I. An Introduction (Springer, 2007).
Giomi, L., Mahadevan, L., Chakraborty, B. & Hagan, M. F. Excitable patterns in active nematics. Phys. Rev. Lett. 106, 218101 (2011).
Woodhouse, F. G. & Goldstein, R. E. Spontaneous circulation of confined active suspensions. Phys. Rev. Lett. 109, 168105 (2012).
Benzi, R. & Ching, E. S. C. Polymers in fluid flows. Annu. Rev. Condens. Matter Phys. 9, 163–181 (2018).
Whitchurch, C. B., Tolker-Nielsen, T., Ragas, P. C. & Mattick, J. S. Extracellular DNA required for bacterial biofilm formation. Science 295, 1487 (2002).
Mukherjee, A., Walker, J., Weyant, K. B. & Schroeder, C. M. Characterization of flavin-based fluorescent proteins: an emerging class of fluorescent reporters. PLoS ONE 8, e64753 (2013).
Mason, T. G., Ganesan, K., van Zanten, J. H., Wirtz, D. & Kuo, S. C. Particle tracking microrheology of complex fluids. Phys. Rev. Lett. 79, 3282–3285 (1997).
Zhu, X., Kundukad, B. & van der Maarel, J. R. Viscoelasticity of entangled λ-phage DNA solutions. J. Chem. Phys. 129, 185103 (2008).
Kundukad, B. & van der Maarel, J. R. C. Control of the flow properties of DNA by topoisomerase II and its targeting inhibitor. Biophys. J. 99, 1906–1915 (2010).
Brochard, F. Viscosities of dilute polymer solutions in nematic liquids. J. Polym. Sci. Polym. Phys. Ed. 17, 1367–1374 (1979).
Acknowledgements
We thank Y. Li and W. Zuo for building the image acquisition and microscope stage temperature control systems, H. C. Berg (Harvard University) for providing the bacterial strains, A. Mukherjee and C. M. Schroeder (UIUC) for providing the pAM06-tet plasmid, and L. Xu (CUHK) for assistance with bulk rheology measurement. We thank E. S.C. Ching (CUHK), K. Xia (CUHK) and T. Ngai (CUHK) for discussions and comments. This work was supported by the National Natural Science Foundation of China (NSFC no. 31971182, to Y.W.), the Research Grants Council of Hong Kong SAR (RGC Ref. No. 14303918 and CUHK Direct Grants; to Y.W.), the US National Science Foundation Grant DMR-1609208 (to M.C.M and S.S) and KITP under grant no. PHY-1748958. S.S. is supported by the Harvard Society of Fellows. M.C.M and S.S thank the KITP for hospitality in the course of this work.
Author information
Authors and Affiliations
Contributions
S.L. discovered the phenomena, designed the study, performed experiments, and analysed and interpreted the data. S.S. and M.C.M. developed the active-matter model, and analysed and interpreted the data. Y.W. conceived the project, designed the study, and analysed and interpreted the data. Y.W. wrote the first draft. All authors contributed to the revision of the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Height profile of bacterial suspension drop and further characterization of giant vortices.
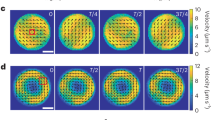
a, Height profile of a bacterial suspension drop measured by locating the uppermost and the lowermost focal planes where fluorescently labelled cells could be found. b, Instantaneous velocity field of a representative CW unidirectional giant vortex. Arrows and colourmap represent collective velocity direction and magnitude, respectively (Methods). Cell density, 6 × 1010 cells ml−1; DNA concentration, 200 ng μl−1. Scale bar, 250 μm. c, Time- and azimuthally averaged tangential velocity vθ of the CW giant vortex in b plotted against radial position. Error bars represent standard deviation (N = 1,000 successive frames). d, Normalized mean vortical flow of the CV giant vortex in b (Methods). e–g, Fourier analysis of the normalized mean vortical flow (P(t)) in oscillatory giant vortices. e, Fourier spectrum |P(f)| for P(t) in Fig. 2b computed by fast Fourier transform, peaking at ~0.030 Hz and with a full-width at half-maximum (FWHM) of ~0.012 Hz. f, P(t) of an oscillatory vortex with nine periods (cell density, ~6 × 10 cells ml−1; DNA concentration, ~800 ng μl−1). g, Fourier spectrum |P(f)| for P(t) in panel f, peaking at ~0.026 Hz and with a FWHM of ~0.008 Hz.
Extended Data Fig. 2 Diffusive behaviour of single cells in giant vortices.
a, The mean square displacement (MSD) of individual cells analysed in Fig. 1e in the laboratory frame (Methods). See more discussion in Methods. b, Bacterial trajectories in Fig. 1e replotted with the starting points shifted to the same position (black dot). Different colour indicates different bacterium. Scale bar, 100 μm. c, Local diffusive behaviour of individual bacteria in an oscillatory giant vortex. MSD of cells was computed based on drift-subtracted single-cell trajectories. In a frame comoving with the giant vortex, individual cells underwent ballistic motion at short timescale (~1 s) and diffusive motion over longer timescales. The diffusion constant D was obtained by fitting the MSD at t > 2 s to \(4D{t}^{\alpha }\), yielding D ≈ 110 μm2 s−1 and α ≈ 1.1. In this oscillatory giant vortex, DNA concentration was ~500 ng μl−1 and cell density was ~6 × 1010 cells ml−1. Inset: trajectories of 14 representative cells (+, starting point; ○, ending point). The time duration of each trajectory is ~28 s, about one period of the oscillatory giant vortex.
Extended Data Fig. 3 Vortex order of bacterial suspension drop versus E. coli DNA concentration.
The diameters of suspension drops were ~1.5 mm. Cell density was fixed at 6 × 10 cells ml−1. a, Scattered data points of vortex order (that is, normalized mean vortical flow P) versus DNA concentration. Each data point represents the normalized mean vortical flow averaged over a time window of ~20 s for one suspension drop with specific DNA concentration. b, Sigmoidal fit of P as a function of DNA concentration. The mean and standard deviation (error bars, N ≥ 4) plotted in b were computed based on the scattered data points in a. The data in b was fitted to a modified sigmoid function (Methods). See more discussion in Methods.
Extended Data Fig. 4 Dependence of mean-vortical-flow amplitude of oscillatory giant vortices on cell density and DNA concentration.
The mean-vortical-flow amplitude of a specific oscillatory giant vortex is taken as the averaged absolute value of extremums of the normalized mean vortical flow. a, Contour plot of mean-vortical-flow amplitude (indicated by the colourmap) in the plane of cell density and DNA concentration. Each data point in the contour plot is the average of mean-vortical-flow amplitude from at least three oscillatory giant vortices with the corresponding DNA concentration and cell density. b, The mean-vortical-flow amplitude in panel a plotted against DNA concentration at fixed cell density ~6 × 10 cells ml−1. c, Mean-vortical-flow amplitude in a plotted against cell density at fixed DNA concentration ~600 ng μl−1. Error bars in b, c indicate standard deviation (N ≥ 3). Overall, the mean-vortical-flow amplitude of oscillatory giant vortices increases weakly with increasing DNA concentration and cell density.
Extended Data Fig. 5 Dynamical states and phase-space trajectories obtained from the theoretical model.
a, The mode structure as a function of τp for fixed α, τR and q ≈ \(\sqrt{|\alpha |/K}\). For τp < τI, we have one purely real unstable mode (Re(σ) > 0), while for τp > τII, the unstable modes have a finite frequency of oscillation. Here σ is the eigenvalue of the linear stability matrix. b, The phase-plane portraits in the \(\{\delta {p}_{\perp },\delta \sigma \}\) plane for the three different regimes τp < τI, τI < τp < τII and τp > τII. We have included the leading gradient-free nonlinear term \(\delta {p}_{\perp }^{3}\) to saturate the polarization when unstable. This makes the system akin to the FitzHugh–Nagumo model for \({\tau }_{{\rm{p}}}\gtrsim {\tau }_{{\rm{II}}}\), leading to relaxation oscillations and excitability. The black and orange lines are the nullclines, and the red line is a representative trajectory that either converges to a fixed point or to a limit cycle. The red stars at the intersection of the nullclines are stable fixed points (or foci), while the blue dots are unstable fixed points (or foci). The labels to the three frames highlight the correspondence between the nature of the dynamical state obtained from the FitzHugh–Nagumo model and the states observed in experiments.
Extended Data Fig. 6 Ratio between active shearing time and DNA relaxation time in giant vortices plotted against DNA concentration.
a–c, The Maxwell relaxation time of DNA solutions τp was measured by microrheology (Methods). The active shearing timescale τa = \(\varGamma {l}_{{\rm{a}}}^{2}/|\alpha |\approx K/{\alpha }^{2}\) in giant vortices cannot be computed precisely, as the relevant parameters are unknown. Instead, τa is estimated as the inverse of shear rate associated with bacterial collective motion, that is, the correlation length of collective velocity field divided by mean collective speed. Cell density was fixed at 4 × 10, 6 × 10 and 8 × 10 cells ml−1 for a, b and c, respectively. The mean and uncertainty of each data point in the plots were computed based on the data of τp and τa measured from at least three giant vortices. Overall, τp approaches τa when unidirectional vortices transit to an oscillatory state, a result qualitatively consistent with our active-matter model.
Extended Data Fig. 7 High-molecular-weight DNA can give rise to giant vortices.
a, DNA concentration threshold (d1) for the transition from bacterial turbulence to unidirectional giant vortex decreases with molecular weight (N) for the three types of DNA tested (E. coli genomic DNA, lambda phage DNA, and salmon testes DNA; Methods). The transition DNA concentration threshold and its uncertainty (indicated by error bars) for different types of DNA molecules was estimated based on sigmoidal fit of the normalized mean vortical flow as a function of DNA concentration; see Methods. b–d, Normalized mean vortical flow of bacterial suspension drop versus E. coli DNA concentration obtained (b, E. coli genomic DNA, same as Extended Data Fig. 3b and replotted here for comparison; c, lambda phage DNA; d, salmon testes DNA). Error bars indicate standard deviation, N ≥ 4. The data in these plots were obtained in the same way as in Extended Data Fig. 3b. The diameters of suspension drops were ~1.5 mm. Cell density was fixed at 6 × 10 cells ml−1. See more discussion in Methods.
Extended Data Fig. 8 Effect of viscosity on bacterial collective motion in suspension drops.
The suspension drop diameter was ~1.5 mm. Cell density was fixed at 6 × 10 cells ml−1. a, b, Mean vortex order and average collective speed of bacterial suspension drops without additive DNA plotted against Ficoll (Ficoll 400, molecular weight 400 kDa; Sigma catalogue number F9378) concentration. The mean vortex order of a specific suspension drop was computed as the time average of absolute instantaneous vortex order (that is, normalized mean vortical flow) over a time window of ~20 s. For a specific Ficoll concentration, the average collective speed of a suspension drop was computed as the time average of collective speed over a time window of ~20 s. c, d, Mean vortex order and average collective speed of bacterial suspension drops with additive DNA plotted against Ficoll concentration. Black (or red) colour indicates the experiments with E. coli genomic DNA concentration 200 (or 800) ng μl−1, which normally supports the development of unidirectional or oscillatory giant vortices, respectively. Neither a stable unidirectional giant vortex nor an oscillatory giant vortex could be observed at all Ficoll concentrations (without additive DNA) or at Ficoll concentrations ≥2.5% (with DNA). Error bars in a–d indicate standard deviation (N ≥ 5 suspension drops).
Extended Data Fig. 9 Dynamic modulus of pure bacterial suspension and DNA solution.
a, Dynamic modulus of pure bacterial suspension measured by a rheometer as a function of frequency (Methods), showing viscoelasticity consistent with the Kelvin–Voigt model. The measurement was made on the scale of ~100 μm, comparable to the length scale of bacterial collective motion. Open circles represent the storage modulus (G′); solid circles represent the loss modulus (G″). The colourmap indicates cell density. The elastic modulus of the bacterial suspension measured in the range of ~0.1–1 Hz was used to compute the data points in Fig. 4e. b, Dynamic modulus of DNA solution measured by microrheology (Methods). The dashed line represents the storage modulus (G′); the solid line represents the loss modulus (G″). The colourmap indicates the DNA concentration. The DNA solution behaves as a Maxwell material. Note that the viscosity η of DNA solutions obtained from our microrheology measurement is much higher than that of water (for example, η ≈ 0.106 Pa s at DNA concentration 200 ng μl−1; Methods). The fact that cells were able to swim at a normal speed of ~20–30 μm s−1 at the DNA concentrations tested here suggest that swimming bacteria induce a strong shear thinning effect in DNA solutions.
Extended Data Fig. 10 Confinement effect on the development of giant vortex state.
a, Without spatial confinement (for example, in centimetre-scale bacterial swarming colonies), dense bacterial active fluids can display collective oscillatory motion with the oscillation frequency independent of cell density as shown in the plot here (error bars indicate standard variation; N = 5). b, c, Oscillation frequency (b) and vortical flow amplitude (c) in oscillatory giant vortices plotted against confinement size (that is, diameter of suspension drops). Each dot in b, c represents the data from one suspension drop with the specified size. Cell density was fixed at ~6 × 10 cells ml−1 and E. coli genomic DNA concentration was fixed at ~300 ng μl−1. d–f, DNA concentration threshold for the transition from bacterial turbulence to unidirectional giant vortex plotted against confinement size in the case of E. coli genomic DNA (d), lambda phage DNA (e) and salmon testes DNA (f). The DNA concentration threshold and its uncertainty (indicated by error bars) were estimated based on sigmoidal fit of normalized mean vortical flow as a function of DNA concentration (Methods). Cell density in d–f was fixed at ~6 × 10 cells ml−1. Taken together, spatial confinement is necessary but not sufficient for giant vortex development; see more discussion in Methods.
Supplementary information
Supplementary Information
This file contains the detailed calculation and discussion of the active matter model, including Supplementary Figures 1-4 and additional references.
Supplementary Video 1
Unidirectional giant vortex. This phase-contrast video shows a stable CCW giant vortex developed in the bacterial suspension drop. The video is played at 60 frames per second with the real elapsed time indicated in the time stamp. It is associated with Fig. 1.
Supplementary Video 2
Oscillatory giant vortex with periodic chirality switching. This phase-contrast shows an oscillatory giant vortex switching its global rotational chirality every ~35 s. The video is played at 60 frames per second with the real elapsed time indicated in the time stamp. It is associated with Fig. 2.
Supplementary Video 3
Collective velocity field of oscillatory giant vortex. This video shows the evolution of velocity field during the chirality switching process. Arrows and colourmap represent collective velocity direction and magnitude, respectively. The video is played at 30 f.p.s. with the real elapsed time indicated in the time stamp. It is associated with Video 2 and Figs. 2, 3. The time label in Fig. 2 is consistent with here, while Time = 0 s in Fig. 3 corresponds to Time = 16.5 s in this video.
Supplementary Video 4
Temporal evolution of the spatial profile of azimuthally averaged tangential velocity in the oscillatory giant vortex. This video is associated with Fig. 2e. The tangential velocity profile oscillates up and down during the chirality switching process, in contrast to the stable tangential velocity profile in the unidirectional giant vortex (Fig. 1c and Extended Data Fig. 1c). The video is played at 60 frames per second with the real elapsed time indicated in the time stamp.
Supplementary Video 5
Numerical solution of the steady state laminar vortex. This video is associated with Supplementary Fig. 3a and shows the time evolution and steady-state profile of the active viscoelastic fluid in a disc. The viscoelastic timescales used are \({\tau }_{{\rm{p}}}/{\tau }_{{\rm{n}}}=1\) and \({\tau }_{{\rm{R}}}/{\tau }_{{\rm{n}}}=1\), which in our simulation corresponds to \({\tau }_{{\rm{I}}} < {\tau }_{{\rm{p}}} < {\tau }_{{\rm{I}}{\rm{I}}}\) and hence a stationary laminar vortex. The three panels show the polarization (top left), the polymer stress (top right) and the flow velocity (bottom left), with the colours representing the respective magnitudes. The black arrows for both p and v capture the respective orientations of polarization and flow, while the black lines in the plot for the stress show the stress anisotropy, their angle being set by \({\tan }^{-1}\left(\frac{{{\rm{\sigma }}}_{{\rm{xx}}}^{{\rm{el}}}-{{\rm{\sigma }}}_{{\rm{yy}}}^{{\rm{el}}}}{{{\rm{\sigma }}}_{{\rm{xy}}}^{{\rm{el}}}}\right)\). In the bottom right corner, the radial profile (r = 0 is the disc center) of both azimuthally averaged tangential (blue) and radial (red) velocity profiles are plotted as a function of time (indicated by the time stamp). While the radial velocity is always close to zero, the tangential velocity settles into a stable shear banded profile denoting an inner vortex surrounded a counter-flowing layer.
Supplementary Video 6
Numerical solution of the oscillating vortex. This video is associated with Fig. 2cd and also Supplementary Fig. 2. It shows the time evolution of the oscillatory dynamics of the confined active viscoelastic fluid in a disc. The viscoelastic timescales used are \({\tau }_{{\rm{p}}}/{\tau }_{{\rm{n}}}=20\) and \({\tau }_{{\rm{R}}}/{\tau }_{{\rm{n}}}=4\), which in our simulation corresponds to \({\tau }_{{\rm{p}}} > {\tau }_{{\rm{I}}{\rm{I}}}\) and hence an oscillating vortex. The three panels as before show the polarization (top left), the polymer stress (top right) and the flow velocity (bottom left), with the colours representing the respective magnitudes, and the black arrows and lines capturing respective orientations. As can be seen, the polarization does not reverse and only exhibits small angle oscillations, while the flow velocity periodically reverses as in the experiment (also shown in snapshots in Supplementary Fig. 2). In the bottom left corner, the radial profile (r = 0 is the disc center) of both azimuthally averaged tangential (blue) and radial (red) velocity profiles are plotted as a function of time (indicated by the time stamp). While the radial velocity is always close to zero, the tangential velocity displays steady oscillations, periodically switching sign (temporal snapshots are plotted in Supplementary Fig. 3b along with a time trace in Supplementary Fig. 3e, also Fig. 2d).
Rights and permissions
About this article
Cite this article
Liu, S., Shankar, S., Marchetti, M.C. et al. Viscoelastic control of spatiotemporal order in bacterial active matter. Nature 590, 80–84 (2021). https://doi.org/10.1038/s41586-020-03168-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-03168-6
This article is cited by
-
Emergent memory from tapping collisions in active granular matter
Communications Physics (2024)
-
Photosynthetically-powered phototactic active nematic liquid crystal fluids and gels
Communications Materials (2024)
-
Self-enhanced mobility enables vortex pattern formation in living matter
Nature (2024)
-
Dynamics of driven Janus particles with pure repulsive interactions over obstacle arrays in active bath
Indian Journal of Physics (2024)
-
Orientational dynamics and rheology of active suspensions in weakly viscoelastic flows
Communications Physics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.