Abstract
Whole-genome doubling (WGD) is common in human cancers, occurring early in tumorigenesis and generating genetically unstable tetraploid cells that fuel tumour development1,2. Cells that undergo WGD (WGD+ cells) must adapt to accommodate their abnormal tetraploid state; however, the nature of these adaptations, and whether they confer vulnerabilities that can be exploited therapeutically, is unclear. Here, using sequencing data from roughly 10,000 primary human cancer samples and essentiality data from approximately 600 cancer cell lines, we show that WGD gives rise to common genetic traits that are accompanied by unique vulnerabilities. We reveal that WGD+ cells are more dependent than WGD− cells on signalling from the spindle-assembly checkpoint, DNA-replication factors and proteasome function. We also identify KIF18A, which encodes a mitotic kinesin protein, as being specifically required for the viability of WGD+ cells. Although KIF18A is largely dispensable for accurate chromosome segregation during mitosis in WGD– cells, its loss induces notable mitotic errors in WGD+ cells, ultimately impairing cell viability. Collectively, our results suggest new strategies for specifically targeting WGD+ cancer cells while sparing the normal, non-transformed WGD− cells that comprise human tissue.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
TCGA data used here are publicly available. All other data supporting this study are available within the article or supplementary figures or are available from the authors upon request.
Code availability
All code used here is available at https://github.com/campbio/Manuscripts/tree/master/Quinton_WGD_2020.
Change history
11 May 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-03591-3
References
Lens, S. M. A. & Medema, R. H. Cytokinesis defects and cancer. Nat. Rev. Cancer 19, 32–45 (2019).
Fujiwara, T. et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437, 1043–1047 (2005).
Ganem, N. J., Godinho, S. A. & Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 460, 278–282 (2009).
Thompson, D. A., Desai, M. M. & Murray, A. W. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr. Biol. 16, 1581–1590 (2006).
Dewhurst, S. M. et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 4, 175–185 (2014).
López, S. et al. Interplay between whole-genome doubling and the accumulation of deleterious alterations in cancer evolution. Nat. Genet. 52, 283–293 (2020).
Selmecki, A. M. et al. Polyploidy can drive rapid adaptation in yeast. Nature 519, 349–352 (2015).
Bielski, C. M. et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat. Genet. 50, 1189–1195 (2018).
Ganem, N. J. et al. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell 158, 833–848 (2014).
Senovilla, L. et al. An immunosurveillance mechanism controls cancer cell ploidy. Science 337, 1678–1684 (2012).
Andreassen, P. R., Lohez, O. D., Lacroix, F. B. & Margolis, R. L. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell 12, 1315–1328 (2001).
Ben-David, U. & Amon, A. Context is everything: aneuploidy in cancer. Nat. Rev. Genet. 21, 44–62 (2019). 10
Storchová, Z. et al. Genome-wide genetic analysis of polyploidy in yeast. Nature 443, 541–547 (2006).
Lin, H. et al. Polyploids require Bik1 for kinetochore-microtubule attachment. J. Cell Biol. 155, 1173–1184 (2001).
Carter, S. L. et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30, 413–421 (2012).
Zack, T. I. et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013).
Antao, N. V., Marcet-Ortega, M., Cifani, P., Kentsis, A. & Foley, E. A. A cancer-associated missense mutation in PP2A-Aα increases centrosome clustering during mitosis. iScience 19, 74–82 (2019).
Maruvka, Y. E. et al. Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat. Biotechnol. 35, 951–959 (2017).
Taylor, A. M. et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell 33, 676–689 (2018).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Broad, D. DepMap Achilles 18Q3 Public https://depmap.org/portal/ (2018).
Godinho, S. A. et al. Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510, 167–171 (2014).
Musacchio, A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 25, R1002–R1018 (2015).
Yang, Z., Lončarek, J., Khodjakov, A. & Rieder, C. L. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 10, 748–751 (2008).
Jemaà, M. et al. Whole-genome duplication increases tumor cell sensitivity to MPS1 inhibition. Oncotarget 7, 885–901 (2016).
Wangsa, D. et al. Near-tetraploid cancer cells show chromosome instability triggered by replication stress and exhibit enhanced invasiveness. FASEB J. 32, 3502–3517 (2018).
Zheng, L. et al. Polyploid cells rewire DNA damage response networks to overcome replication stress-induced barriers for tumour progression. Nat. Commun. 3, 815 (2012).
Santaguida, S. & Amon, A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat. Rev. Mol. Cell Biol. 16, 473–485 (2015).
Stumpff, J., von Dassow, G., Wagenbach, M., Asbury, C. & Wordeman, L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev. Cell 14, 252–262 (2008).
Stumpff, J., Wagenbach, M., Franck, A., Asbury, C. L. & Wordeman, L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev. Cell 22, 1017–1029 (2012).
Fonseca, C. L. et al. Mitotic chromosome alignment ensures mitotic fidelity by promoting interchromosomal compaction during anaphase. J. Cell Biol. 218, 1148–1163 (2019).
Mayr, M. I. et al. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr. Biol. 17, 488–498 (2007).
Czechanski, A. et al. Kif18a is specifically required for mitotic progression during germ line development. Dev. Biol. 402, 253–262 (2015).
Hatch, E. M., Fischer, A. H., Deerinck, T. J. & Hetzer, M. W. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154, 47–60 (2013).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Zhang, C.-Z. et al. Chromothripsis from DNA damage in micronuclei. Nature 522, 179–184 (2015).
Janssen, L. M. E. et al. Loss of Kif18A results in spindle assembly checkpoint activation at microtubule-attached kinetochores. Curr. Biol. 28, 2685–2696 (2018).
Edzuka, T. & Goshima, G. Drosophila kinesin-8 stabilizes the kinetochore-microtubule interaction. J. Cell Biol. 218, 474–488 (2019).
Cohen-Sharir, Y. et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature https://doi.org/10.1038/s41586-020-03114-6 (2021).
Marquis, C. et al. Chromosomally unstable tumor cells specifically require KIF18A for proliferation. Nat. Comm. (in the press).
Zhu, H. et al. Targeted deletion of Kif18a protects from colitis-associated colorectal (CAC) tumors in mice through impairing Akt phosphorylation. Biochem. Biophys. Res. Commun. 438, 97–102 (2013).
Zhang, C. et al. Kif18A is involved in human breast carcinogenesis. Carcinogenesis 31, 1676–1684 (2010).
Ellrott, K. et al. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 6, 271–281 (2018).
Knijnenburg, T. A. et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 23, 239–254 (2018).
Bailey, M. H. et al. Comprehensive characterization of cancer driver genes and mutations. Cell 173, 371–385 (2018).
Thorsson, V. et al. The immune landscape of cancer. Immunity 48, 812–830 (2018).
Miao, D. et al. Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat. Genet. 50, 1271–1281 (2018).
Liu, D. et al. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat. Med. 25, 1916–1927 (2019).
Mermel, C. H., Schumacher, S. E., Hill, B., Meyerson, M. L., Beroukhim, R. & Getz, G. GISTIC20 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011).
Federico, A. & Monti, S. hypeR: an R package for geneset enrichment workflows. Bioinformatics 36, 1307–1308 (2020).
McFarland, J. M. et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 9, 4610 (2018).
Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet. 49, 1779–1784 (2017).
Ben-David, U. et al. Patient-derived xenografts undergo mouse-specific tumor evolution. Nat. Genet. 49, 1567–1575 (2017).
Acknowledgements
The findings published here are in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). We thank J. Weinberg for statistical advice; J. Stumpff for technical advice, reagents, and sharing unpublished data; D. Pellman for sharing reagents; E. Van Allen and D. Liu for technical advice; and S. Carter for sharing ABSOLUTE data for cell lines in the CCLE. R.J.Q. is supported by a Canadian Institutes of Health Research Doctoral Foreign Study Award (152266). N.J.G. is a member of the Shamim and Ashraf Dahod Breast Cancer Research Laboratories and is supported by National Institutes of Health (NIH) grants CA154531 and GM117150, the Karin Grunebaum Foundation, the Smith Family Awards Program, the Melanoma Research Alliance and the Searle Scholars Program. This work was also supported in part by a pilot grant from the American Cancer Society (ACS) and the Boston University Clinical and Translational Science Institute Bioinformatics Group, who are in turn supported by a grant from the NIH/National Center for Advancing Translational Sciences (NCATS; 1UL1TR001430).
Author information
Authors and Affiliations
Contributions
R.J.Q. and N.J.G. designed the experiments and wrote the manuscript. R.J.Q. performed most of the cell biological assays and imaging analysis. A.D., C.J.T. and S.P. assisted R.J.Q. with the cell biological assays. K.K. and M.A.V. analysed images. T.S.K. generated the isogenic diploid and tetraploid HCT-116 cells. J.V. generated cell lines. N.H. and A.L.M. performed animal studies. R.J.Q., A.M.T., Y.K., N.P. and J.D.C. performed the computational analyses. All authors edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Mutational burden in WGD+ and WGD− tumours.
a, Total mutational burden in the indicated tumour subtypes across 9,240 TCGA samples (horizontal dotted lines show medians; two-sided Wilcoxon rank-sum test; red asterisks indicate higher burden in WGD− samples and blue asterisks indicate higher burden in WGD+ samples). b, Ploidy-corrected mutational burden in the indicated tumour subtypes across 9,240 TCGA samples (dotted lines show medians; two-sided Wilcoxon rank-sum test; red asterisks indicate higher burden in WGD− samples and blue asterisks indicate higher burden in WGD+ samples). c, Ploidy-corrected mutational burden in WGD+ and WGD− samples in the TCGA (n = 9,414 samples; dotted line shows means ± s.d.). d, Ploidy-corrected mutational burden of TCGA WGD+ and WGD− samples with MSI/POLE mutations (n = 174 samples). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 2 Characteristics of WGD+ cells.
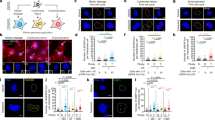
a, Correlation of the fraction of stromal cells in the indicated tumour subtypes with WGD (Pearson’s correlation). b, Correlation of purity with WGD (Pearson’s correlation). c, Illustration of our PSL analysis using gene essentiality scores for KIF18A in the Project Achilles CRISPR data set. Asterisked P values in blue represent instances where the cutoff for enrichment in WGD+ cell lines was met in either our thresholded (two-sided Fisher’s exact) or our non-thresholded (two-sided Wilcoxon’s) analyses (see Methods). CNS, central nervous system. d, Fractions of individuals whose cancers responded or did not respond to blockade of PD1, according to WGD status (two-sided Fisher’s exact test; P = 0.0351). See refs. 48,49. e, Rate of chromosome missegregation in HCT116 cells (n = 3,107 2N cells, 2,594 4N cells; shown are means ± s.d.). f, DNA fluorescence-activated cell sorting (FACS) profile of diploid and tetraploid HCT116 cells at 40 and 70 days of culture. g, Karyotype of diploid and tetraploid HCT-116 cells with the modal chromosome number and range (n = 20 karyotypes analysed per condition). h, Previously published data3,22 showing the stability of isogenic diploid and tetraploid RPE and MCF10A cell lines. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 3 Validation of WGD+ vulnerabilities in isogenic 2N/4N cells.
a, Mitotic duration of the indicated cells following the indicated treatments (n = 200 cells; two-sided Student’s unpaired t-test; shown are means ± s.e.m.; P < 0.0001, < 0.0001, 0.0265, respectively). b, Fractions of mitoses that generate micronuclei following the indicated treatments (n = 200 cells; two-sided Student’s unpaired t-test; P < 0.0001, < 0.0001, < 0.0001, < 0.0001, 0.0002, < 0.0001, respectively). c, Left, relative viability of 2N and 4N HCT116 cells seven days after treatment with the indicated siRNAs at the indicated concentrations. Right, western blot showing protein knockdown 48 h after treatment with siRNA (n = 3 independent experiments; shown are means ± s.e.m. at each dose; for gel source data, see Supplementary Fig. 1). d, Relative viability of 2N and 4N MCF10A cells seven days after treatment with the indicated siRNAs at 50 pM concentration (n = 3 independent experiments; one-sided Student’s unpaired t-test; means ± s.e.m.; P < 0.0001, < 0.0001). e, Relative viability of 2N and 4N RPE cells five days after treatment with the indicated siRNAs at 50 pM concentration (n = 3 independent experiments; one-sided Student’s unpaired t-test; means ± s.e.m.; P = 0.090, 0.0007, respectively). f, Representative western blot showing knockdown of the indicated proteins 48 h after treatment with the indicated siRNAs (n = 3 independent experiments; for gel source data, see Supplementary Fig. 1). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 4 Validation of WGD+ vulnerabilities in breast-cancer cells.
a, b, Dose–response curves for the indicated cell lines following the indicated treatments after seven days, with accompanying LC50 values (n = 3 independent experiments; nonlinear regression with variable slope; graphs show mean relative viability ± s.e.m. at each dose or mean LC50 ± 95% confidence interval). c–e, Dose–response curves for 5 WGD− and 5 WGD+ breast cancer cell lines seven days after treatment with the indicated drugs and concentrations (n = 3 independent experiments; nonlinear regression with variable slope; means ± s.e.m. at each dose). f, Representative western blot showing knockdown of the indicated proteins in breast-cancer cell lines 48 h after treatment with the indicated siRNAs (n = 3 independent experiments; for gel source see Supplementary Fig. 1). g, Relative viability decrease in WGD+ and WGD− breast-cancer cell lines seven days after treatment with the indicated siRNAs (two-sided Wilcoxon’s rank-sum test; means ± s.e.m.; P < 0.0001 and P = 0.0027, respectively). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 5 Mitotic fidelity in WGD+ cells following KIF18A depletion.
a, Dose–response curves for the indicated cell lines following a seven-day treatment with MG132, with accompanying LC50 values (n = 3 independent experiments; nonlinear regression with variable slope; graphs show mean relative viability ± s.e.m. at each dose and mean LC50 ± 95% confidence interval). b, Progression-free survival and overall survival for individuals in the upper tertile of tumour KIF18A expression in the TCGA (Cox’s proportional-hazards regression; graph shows hazard ratios ± 95% confidence interval). c, Representative western blot showing KIF18A levels following transfection with the indicated siRNAs in the indicated cell lines (n = 3 independent experiments; for gel source data, see Supplementary Fig. 1). d, Anaphase phenotypes following depletion of KIF18A (n = 20 cells per condition; asterisks indicate P values from two-sided Fisher’s exact tests comparing the fraction of anaphases with lagging chromosomes; P < 0.0001, 0.0033, 0.0187, respectively). e, Representative confocal images showing phases of mitosis in the indicated cell lines 48 h after transfection with the indicated siRNAs (representative images from two independent experiments; scale bar, 10 μm). f, Representative still images from 2N and 4N MCF10A cells progressing through mitosis after transfection with the indicated siRNAs. Chromosomes labelled with H2B–GFP are shown in white. Arrows in enlarged images show oscillating chromosomes during metaphase and the generation of a micronucleus (shown are h:min; scale bar, 10 μm). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 6 Analysis of cell fate in WGD+ cells following KIF18A depletion.
a, Left, representative images of a 4N MCF10A cell four days after transfection with siKIF18A and stained for DNA, cGAS and γ-H2AX. Right, fraction of micronuclei in 2N and 4N MCF10A cells with the indicated treatments that stained positive for cGAS (n = 200 micronuclei per condition; two-sided Fisher’s exact test; scale bar, 10 μm; P <0.0001, P = 0.0069, respectively). b, c, Representative confocal images of the indicated cell lines 48 h after transfection with the indicated siRNAs. Arrowheads highlight MAD1-positive kinetochores in misaligned chromosomes (scale bar, 10 μm; representative images from two independent experiments). d, Right, representative western blot showing levels of the indicated proteins after treatment with the indicated siRNAs, and left, graphs showing relative protein levels normalized to loading control (n = 3 independent experiments; one-sided Student’s unpaired t-test; means ± s.e.m.; P = 0.0337, 0.0030, 0.0674, 0.0421, 0.0067, 0.0227, respectively; for gel source data, see Supplementary Fig. 1). e, Cell fates of the indicated lines, tracked for 3 days beginning 18 h after transfection with the indicated siRNAs (n = 40 cells per condition; two-sided Fisher’s exact test comparing the fraction of cells arresting/delaying in interphase relative to the control group; P = 0.0016, < 0.0001, < 0.0001, respectively). f, Relative viability of the indicated cell lines four days after transfection with the indicated siRNAs (n = 3 independent experiments; two-sided Student’s unpaired t-test; means ± s.e.m.; P = 0.0132, 0.0310, 0.8808, 0.8615, respectively). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 7 PSL effect of KIF18A depletion.
a, Bottom left, western blot showing endogenous KIF18A levels in the indicated cell lines; top and right, graphs showing respective protein levels normalized to the GAPDH loading control (representative blot from three independent experiments; for gel source data, see Supplementary Fig. 1). b, Representative western blot showing KIF18A levels 48 h after transfection with the indicated siRNAs (n = 3 independent experiments; for gel source data, see Supplementary Fig. 1). c, Relative viability decrease in WGD+ and WGD− breast-cancer cell lines seven days after treatment with the indicated siRNAs (n = 3 independent experiments; two-sided Wilcoxon’s rank-sum test; means ± s.e.m.; P < 0.0001). d, Right, relative viability seven days after induction of Cas9 in cells with sgRNA targeting KIF18A. Left, western blot showing protein depletion 72 h after induction (n = 3 independent experiments; means ± s.e.m.; two-sided Student’s unpaired t-test; P = 0.0007, < 0.0001, respectively; for gel source data, see Supplementary Fig. 1). e, Relative viability seven days after induction of shRNA targeting KIF18A, with a western blot showing protein depletion 120 h after induction (n = 3 independent experiments; means ± s.e.m.; one-sided Student’s unpaired t-test; P < 0.0001; for gel source data, see Supplementary Fig. 1). f, Chromosomes with the widest oscillations in each poleward direction immediately before anaphase (n = 20 cells per condition; two-sided Student’s unpaired t-test; P = 0.0022, 0.1781, 0.1487, 0.0136, 0.0820, < 0.0001, < 0.0001, < 0.0001, < 0.0001, 0.4132, respectively). g, Two-dimensional cross-sectional area of the entire body of chromosomes immediately before anaphase (n = 20 cells per condition; two-sided Student’s unpaired t-test; P = 0.1178, 0.7545, 0.1440, 0.0034, 0.9989, 0.0005, 0.0033, 0.0012, 0.0110, 0.9089, respectively; means ± s.e.m.). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Extended Data Fig. 8 Effects of KIF18 depletion in aneuploid cells.
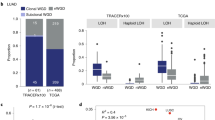
a, Measurement of spindle length (centrosome-to-centrosome) after transfection with the indicated siRNAs (n = 20 cells per condition; two-sided Student’s unpaired t-test; shown are means ± s.e.m.). b, Anaphase phenotypes following depletion of KIF18A (n = 20 cells per condition; asterisks indicate P values from two-sided Fisher’s exact tests comparing the fraction of anaphases with lagging chromosomes). c, Fractions of cells in each cell line undergoing the indicated fates after completing a KIF18A-deficient mitosis that resulted in the formation of micronuclei (n = 25 cells per condition). d, Fractions of cells in each cell line that experience mitotic death in their first and second mitoses following KIF18A depletion (n = 25 cells per condition). e, KIF18A essentiality scores for WGD− and WGD+ cell lines, segregated into ‘highly aneuploid’ (aneuploidy score (AS) score of 10 or more) and ‘non-highly aneuploid’ (AS < 10) categories on the basis of aneuploidy scores (see Methods) (dotted lines show means; two-sided Wilcoxon’s rank-sum test; P = 0.02583, 0.3682, respectively). f, Aneuploidy scores and WGD status for 998 cancer cell lines in the CCLE. g, Relative viability of the indicated cell lines seven days after transfection with the indicated siRNAs (n = 3 independent experiments; each condition normalized to respective control; one-way ANOVA with Dunnett’s post hoc test; means ± s.e.m.; P = 0.1676, > 0.9999, 0.0040, 0.2698, 0.0007, respectively). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Supplementary information
Supplementary Figure 1
Original western blots for all experiments in the manuscript. All images are uncropped, and molecular weight markers are labeled. Corresponding loading controls are indicated.
Supplementary Table 1
Pan-cancer gene expression analysis with GSEA and gene expression analysis for each tumor subtype for WGD+ tumors in the TCGA.
Supplementary Table 2
ABSOLUTE algorithm applied to cancer cell lines indicating purity, ploidy, and number of whole genome doublings.
Supplementary Table 3
Gene essentiality data for ~600 cancer cell lines from the Cancer Cell Line Encyclopedia showing genes enriched for essentiality in WGD+ cell lines.
Supplementary Table 4
List of ploidy-specific lethal genes ranked by their PSL score.
Supplementary Table 5
Cell lines from the CCLE with accompanying aneuploidy scores.
Video 1
Live-cell imaging of diploid (2N) MCF10A H2B-GFP cells following transfection with control siRNA (5 frames/second; hour:minute; scale bar 10 μm).
Video 2
Live-cell imaging of diploid (2N) MCF10A H2B-GFP cells following transfection with KIF18A siRNA (5 frames/second; hour:minute; scale bar 10 μm).
Video 3
Live-cell imaging of tetraploid (4N) MCF10A H2B-GFP cells following transfection with control siRNA (5 frames/second; hour:minute; scale bar 10 μm).
Video 4
Live-cell imaging of tetraploid (4N) MCF10A H2B-GFP cells following transfection with KIF18A siRNA (5 frames/second; hour:minute; scale bar 10 μm).
Video 5
Live-cell imaging of the HCC1806 H2B-GFP breast cancer cell line following transfection with control siRNA (40 frames/second; hour:minute; scale bar 100 μm).
Video 6
Live-cell imaging of the HCC1806 H2B-GFP breast cancer cell line following transfection with KIF18A siRNA (40 frames/second; hour:minute; scale bar 100 μm).
Rights and permissions
About this article
Cite this article
Quinton, R.J., DiDomizio, A., Vittoria, M.A. et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 590, 492–497 (2021). https://doi.org/10.1038/s41586-020-03133-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-020-03133-3
This article is cited by
-
scAbsolute: measuring single-cell ploidy and replication status
Genome Biology (2024)
-
Unifying duplication episode clustering and gene-species mapping inference
Algorithms for Molecular Biology (2024)
-
Aneuploid serves as a prognostic marker and favors immunosuppressive microenvironment in ovarian cancer
Journal of Ovarian Research (2024)
-
Tumor acidosis-induced DNA damage response and tetraploidy enhance sensitivity to ATM and ATR inhibitors
EMBO Reports (2024)
-
Therapeutic targeting of the KIF18A motor protein in cancers with chromosomal instability
Nature Cancer (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.