Abstract
Pyrethroid-impregnated bed nets have driven considerable reductions in malaria-associated morbidity and mortality in Africa since the beginning of the century1. The intense selection pressure exerted by bed nets has precipitated widespread and escalating resistance to pyrethroids in African Anopheles populations, threatening to reverse the gains that been made by malaria control2. Here we show that expression of a sensory appendage protein (SAP2), which is enriched in the legs, confers pyrethroid resistance to Anopheles gambiae. Expression of SAP2 is increased in insecticide-resistant populations and is further induced after the mosquito comes into contact with pyrethroids. SAP2 silencing fully restores mortality of the mosquitoes, whereas SAP2 overexpression results in increased resistance, probably owing to high-affinity binding of SAP2 to pyrethroid insecticides. Mining of genome sequence data reveals a selective sweep near the SAP2 locus in the mosquito populations of three West African countries (Cameroon, Guinea and Burkina Faso) with the observed increase in haplotype-associated single-nucleotide polymorphisms mirroring the increasing resistance of mosquitoes to pyrethroids reported in Burkina Faso. Our study identifies a previously undescribed mechanism of insecticide resistance that is likely to be highly relevant to malaria control efforts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
References
Bhatt, S. et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature 526, 207–211 (2015).
WHO. World Malaria Report (WHO, 2018).
Hawley, W. A. et al. Community-wide effects of permethrin-treated bed nets on child mortality and malaria morbidity in western Kenya. Am. J. Trop. Med. Hyg. 68, 121–127 (2003).
Churcher, T. S., Lissenden, N., Griffin, J. T., Worrall, E. & Ranson, H. The impact of pyrethroid resistance on the efficacy and effectiveness of bednets for malaria control in Africa. eLife 5, e16090 (2016).
Protopopoff, N. et al. Effectiveness of a long-lasting piperonyl butoxide-treated insecticidal net and indoor residual spray interventions, separately and together, against malaria transmitted by pyrethroid-resistant mosquitoes: a cluster, randomised controlled, two-by-two factorial design trial. Lancet 391, 1577–1588 (2018).
Stevenson, B. J. et al. Cytochrome P450 6M2 from the malaria vector Anopheles gambiae metabolizes pyrethroids: sequential metabolism of deltamethrin revealed. Insect Biochem. Mol. Biol. 41, 492–502 (2011).
Müller, P. et al. Field-caught permethrin-resistant Anopheles gambiae overexpress CYP6P3, a P450 that metabolises pyrethroids. PLoS Genet. 4, e1000286 (2008).
Gleave, K., Lissenden, N., Richardson, M. & Ranson, H. Piperonyl butoxide (PBO) combined with pyrethroids in long-lasting insecticidal nets (LLINs) to prevent malaria in Africa. Cochrane Database Syst. Rev. 8, CD012776 (2017).
Toe, K. H. et al. Do bednets including piperonyl butoxide offer additional protection against populations of Anopheles gambiae s.l. that are highly resistant to pyrethroids? An experimental hut evaluation in Burkina Fasov. Med. Vet. Entomol. 32, 407–416 (2018).
Ingham, V. A., Wagstaff, S. & Ranson, H. Transcriptomic meta-signatures identified in Anopheles gambiae populations reveal previously undetected insecticide resistance mechanisms. Nat. Commun. 9, 5282 (2018).
Edi, C. V. et al. CYP6 P450 enzymes and ACE-1 duplication produce extreme and multiple insecticide resistance in the malaria mosquito Anopheles gambiae. PLoS Genet. 10, e1004236 (2014).
Vieira, F. G. & Rozas, J. Comparative genomics of the odorant-binding and chemosensory protein gene families across the Arthropoda: origin and evolutionary history of the chemosensory system. Genome Biol. Evol. 3, 476–490 (2011).
Leal, W. S. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58, 373–391 (2013).
Iovinella, I., Bozza, F., Caputo, B., Della Torre, A. & Pelosi, P. Ligand-binding study of Anopheles gambiae chemosensory proteins. Chem. Senses 38, 409–419 (2013).
Xuan, N. et al. Increased expression of CSP and CYP genes in adult silkworm females exposed to avermectins. Insect Sci. 22, 203–219 (2015).
Bautista, M. A. M. et al. Evidence for trade-offs in detoxification and chemosensation gene signatures in Plutella xylostella. Pest Manag. Sci. 71, 423–432 (2015).
Liu, G. X. et al. Biotype expression and insecticide response of Bemisia tabaci chemosensory protein-1. Arch. Insect Biochem. Physiol. 85, 137–151 (2014).
Yunta, C. et al. Pyriproxyfen is metabolized by P450s associated with pyrethroid resistance in An. gambiae. Insect Biochem. Mol. Biol. 78, 50–57 (2016).
The Anopheles gambiae 1000 Genomes Consortium. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552, 96–100 (2017).
Voight, B. F., Kudaravalli, S., Wen, X. & Pritchard, J. K. A map of recent positive selection in the human genome. PLoS Biol. 4, e72 (2006).
Sabeti, P. C. et al. Genome-wide detection and characterization of positive selection in human populations. Nature 449, 913–918 (2007).
Pombi, M. et al. Unexpectedly high Plasmodium sporozoite rate associated with low human blood index in Anopheles coluzzii from a LLIN-protected village in Burkina Faso. Sci. Rep. 8, 12806 (2018).
Namountougou, M. et al. Multiple insecticide resistance in Anopheles gambiae s.l. populations from Burkina Faso, West Africa. PLoS ONE 7, e48412 (2012).
Harris, C. et al. Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog. 6, e1001112 (2010).
Adolfi, A., Pondeville, E., Lynd, A., Bourgouin, C. & Lycett, G. J. Multi-tissue GAL4-mediated gene expression in all Anopheles gambiae life stages using an endogenous polyubiquitin promoter. Insect Biochem. Mol. Biol. 96, 1–9 (2018).
WHO. Test Procedures for Insecticide Resistance Monitoring in Malaria Vector Mosquitoes (WHO, 2016).
Severo, M. S. et al. Unbiased classification of mosquito blood cells by single-cell genomics and high-content imaging. Proc. Natl Acad. Sci. USA 115, E7568–E7577 (2018).
Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C t method. Nat. Protocols 3, 1101–1108 (2008).
Sockolosky, J. T. & Szoka, F. C. Periplasmic production via the pET expression system of soluble, bioactive human growth hormone. Protein Expr. Purif. 87, 129–135 (2013).
Santolamazza, F. et al. Insertion polymorphisms of SINE200 retrotransposons within speciation islands of Anopheles gambiae molecular forms. Malar. J. 7, 163 (2008).
Giraldo-Calderón, G. I. et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 43, D707–D713 (2015).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Carpenter, B. et al. Stan: a probabilistic programming language. J. Stat. Softw. 76, 1–32 (2017).
Acknowledgements
We thank N. Grisales (Bakaridjan in 2013 and 2015), A. Sanou, M. Guelbeogo (Tiefora 2018, Tengrela 2011, 2012, 2014, 2016) and N. Lissenden (Tengrela 2018) for providing field collections; D. Neafsey and J. Tennessen for sharing Tengrela whole-genome sequence data; I. Iovinella for the provision of SAP1 and SAP3 plasmids; M. Bernardi for help with the generation of the map in Fig. 3a and the preparation of figures; F. Brown, S. Elg, P. Pignatelli and D. Au for providing technical support; H. Toé, B. Lambert and T. Churcher for supplying both the field data and figure in Extended Data Fig. 8; and D. Tsakireli and E. Morou for their help with the expression and characterization of SAP proteins. This study was funded by an MRC Skills Development Fellowship (MR/R024839/1) to V.A.I. and a Royal Society Challenge Grant (CH160059) to H.R. Mosquito collections in Burkina Faso were supported by EC FP7 Project grant no: 265660 ‘AvecNet’ and Wellcome Trust Collaborative Award (200222/Z/15/Z).
Author information
Authors and Affiliations
Contributions
V.A.I. and H.R. conceived the experimental design. V.A.I. performed all transcriptomic expression experiments, RNAi and phenotyping experiments, data analysis, and PCR and associated sequencing experiments. A.A. and G.L. produced the transgenic lines and associated phenotypic characterisation. V.D. and J.V. performed the binding assays and associated protein expression experiments. N.J.H. analysed the data from the Anopheles gambiae 1000 Genomes Project and produced the haplotype SNP panel. M.M. provided all insectary support. V.A.I. and H.R. drafted the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature thanks Mara Lawniczak, James G Logan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Chemosensory protein cluster.
a, Schematic of the region surrounding the shared haplotype block found in the Anopheles gambiae 1000 Genomes Project data with all chemosensory proteins in the cluster highlighted in yellow. Genes displayed in order of appearance, from left to right, are as follows: AGAP008046, AGAP013713, AGAP008047, AGAP008048, AGAP008049, AGAP008050, AGAP008051 (SAP1), AGAP008052 (SAP2), AGAP008053, AGAP008054 (SAP3), AGAP008055 (CSP3), AGAP008056, AGAP029127 (CSP5, previously AGAP008058), AGAP008059 (CSP1), AGAP008060, AGAP008061 and AGAP008062 (CSP4). b, cDNA bootstrap consensus tree inferred from 1,000 replicates using the maximum likelihood method; the percentage of replicate trees with the associated clustering are shown next to the branches. Yellow indicates the sensory appendage proteins, orange the remaining chemosensory proteins in the 3R cluster and black dotted lines show CSP6, which is located on 2R. Alternative isoforms are represented with ‘-RX’, with ‘X’ proceeding alphabetically dependent on number of splice variants.
Extended Data Fig. 2 Overexpression of the CSP family in a multi-insecticide-resistant Anopheles population.
Left, mean relative fold change of each CSP in Tiassalé mosquitoes (blue) compared with the susceptible control N’Gousso mosquitoes (grey) as determined by qPCR. Right, mean relative fold change of each CSP in Tiassalé mosquitoes (blue) compared with the susceptible Kisumu population (grey). Points show three biological replicates. Data are mean ± s.d. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Statistical significance was calculated by an ANOVA followed by Dunnett’s post hoc test; P values are included in Supplementary Table 2.
Extended Data Fig. 3 Expression levels of non-induced chemosensory proteins after exposure to deltamethrin in Tiassalé mosquitoes.
a, Expression levels of the remaining four CSPs at various time points after exposure to deltamethrin in the multi-insecticide-resistant Tiassalé population. b, Tissue-specific induction of these four CSPs 4-h after exposure to deltamethrin. Data are mean ± s.d. of three biological replicates. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Statistical significance was calculated by an ANOVA followed by Dunnett’s post hoc test. If the data were non-normal, data were analysed using a Kruskall–Wallis test followed by a Dunn’s post hoc test; P values are included in Supplementary Table 2.
Extended Data Fig. 4 Efficacy of RNAi.
mRNA knockdown of whole female mosquitoes 72 h after injection compared with GFP-injected controls. Data are mean ± s.d. of three biological replicates.
Extended Data Fig. 5 Phenotype of other induced CSPs to a panel of insecticides.
a, Effect of attenuation of dsSAP3 on mortality after insecticide exposure in Tiassalé mosquitoes (right bars; deltamethrin, n = 4; permethrin, n = 5; α-cypermethrin, n = 3; DDT, n = 3; pirimiphos-methyl, n = 3; bendiocarb, n = 4) compared with dsGFP-injected controls (left bars, green patterned; deltamethrin, n = 5; permethrin, n = 5; α-cypermethrin, n = 5; DDT, n = 4; pirimiphos-methyl, n = 4; bendiocarb, n = 8). b, Effect of attenuation of dsCSP4 on mortality after insecticide exposure in Tiassalé mosquitoes (right bars; deltamethrin, n = 3; permethrin, n = 3; α-cypermethrin, n = 3; DDT, n = 3; pirimiphos-methyl, n = 3; bendiocarb, n = 3) compared with dsGFP-injected controls (left bars, green patterned; deltamethrin, n = 5; permethrin, n = 5; α-cypermethrin, n = 5; DDT, n = 4; pirimiphos-methyl, n = 4; bendiocarb, n = 8). c, Effect of attenuation of dsCSP6 on mortality after insecticide exposure in Tiassalé mosquitoes (right bars; deltamethrin, n = 6; permethrin, n = 4; α-cypermethrin, n = 4; DDT, n = 3; pirimiphos-methyl, n = 4; bendiocarb, n = 5) compared with dsGFP-injected controls (left bars, green patterned; deltamethrin, n = 5; permethrin, n = 5; α-cypermethrin, n = 5; DDT, n = 4; pirimiphos-methyl, n = 4; bendiocarb, n = 8). Analysis of mortality data was done using an ANOVA followed by a Tukey post hoc test; n.s indicates a non-significant change in mortality; *P ≤ 0.05. dsCSP6 μmortality = 11.7–31.6%, P = 0.0474. N indicates the number of individual mosquitoes used for phenotyping; points show the number of bioassay replicates per group. Data are mean ± s.d.
Extended Data Fig. 6 Characterization of SAP2 in the transgenic line.
a, Mean mRNA expression after SAP2 overexpression in the SAP2 × A10 transgenic line (n = 2) compared with SAP2 expression in the A10 × G3 control (n = 3). Data are mean ± s.d. and points show each biological replicate. b, mCherry under the PUBc A10 promoter demonstrating (i) ubiquitous expression; (ii) expression in the head; and (iii) expression in the legs as previously shown25; these results were tested across more than 100 independent mosquito screens. c, Intron splicing confirmed by PCR in A10 × SAP2 and negative control A10 mosquitoes compared with plasmid DNA of pUAS:SAP2. The size of the PCR product with and without the synthetic intron is 647 bp and 534 bp, respectively. MW, 100-bp DNA ladder. n = 2 A10 × SAP2 samples and n = 2 A10 control samples (each sample consists of a pool of 5 4-day-old, unfed females) were tested and experiments were repeated in 2 PCRs.
Extended Data Fig. 7 Effect of SAP2 RNAi injection on the fitness of Tiassalé mosquitoes.
a, Longevity of SAP2 RNAi (dsSAP2)-expressing mosquitoes (black) compared with dsGFP-expressing control mosquitoes (green). N indicates the number of individual mosquitoes used in each group; n.s represents P = 0.113 as calculated by a two-sided Mantel–Cox test. b, Life-history traits of dsSAP2-injected (black) and dsGFP-injected (green) females. (i) Number of eggs in each group 72-h after a blood meal (the median and interquartile range are shown). (ii) Proportion of females with eggs (dark shading indicates females with eggs, light shading without; P = 0.4382). (iii) Mortality after a blood meal (dark shading are females that are alive after a blood meal, light those that are dead; P = 0.0052). (iv) Blood feeding proportions (dark shading are blood-fed females, light non-blood-fed; P = 0.3257). Numbers show total numbers of individual females in each group. Significance in (i) was calculated by a two-tailed Mann–Whitney U-test (n.s represents P = 0.0657); significance in (ii), (iii) and (iv) was calculated using a χ2 test. **P ≤ 0.01.
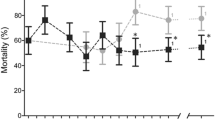
Extended Data Fig. 8 Mortality of A. coluzzii field populations.
Temporal plot of mortality from 2011 to 2018 of A. coluzzii mosquitoes to 0.05% WHO tube deltamethrin exposure. Δ is the posterior median change in mortality from 2011 to 2018. N indicates the number of experiments included (minimum sample size for any given data point is 14). p indicates the posterior probability that resistance (the proportion of posterior samples for which the April 2018 mean exceeds the corresponding value in January 2011) has increased over the time period. The blue line indicates the posterior median of a logistic model fit to binomial test results; the two parameters of the logistic function were assigned using uninformative (Cauchy(0, 1)) priors. The model was fitted using Stan33 with 4 chains and 800 iterations per chain (400 of which were discarded as burn-in in each case); all parameters had Rhat < 1.1, indicating convergence. The shading indicates the 90% predictive interval on the mean. Data and figure were provided by H. Toé, B. Lambert and T. Churcher.
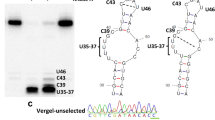
Extended Data Fig. 9 Sequencing of SAP2 primer binding sites.
Four N’Gousso and four Tiassale mosquitoes were sequenced across the primer binding sites. a, Complete conservation of the sequence was seen in the forward binding site. b, One N’Gousso mosquito was heterozygous at one base in the centre of the reverse primer binding site.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-3.
Source data
Rights and permissions
About this article
Cite this article
Ingham, V.A., Anthousi, A., Douris, V. et al. A sensory appendage protein protects malaria vectors from pyrethroids. Nature 577, 376–380 (2020). https://doi.org/10.1038/s41586-019-1864-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1864-1
This article is cited by
-
The impact of agrochemical pollutant mixtures on the selection of insecticide resistance in the malaria vector Anopheles gambiae: insights from experimental evolution and transcriptomics
Malaria Journal (2024)
-
Overcoming insecticide resistance in Anopheles mosquitoes by using faster-acting solid forms of deltamethrin
Malaria Journal (2023)
-
The interplay between malaria vectors and human activity accounts for high residual malaria transmission in a Burkina Faso village with universal ITN coverage
Parasites & Vectors (2023)
-
Deltamethrin and transfluthrin select for distinct transcriptomic responses in the malaria vector Anopheles gambiae
Malaria Journal (2023)
-
Evaluation of diffuse reflectance spectroscopy for predicting age, species, and cuticular resistance of Anopheles gambiae s.l under laboratory conditions
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.