Abstract
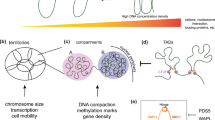
Features of higher-order chromatin organization—such as A/B compartments, topologically associating domains and chromatin loops—are temporarily disrupted during mitosis1,2. Because these structures are thought to influence gene regulation, it is important to understand how they are re-established after mitosis. Here we examine the dynamics of chromosome reorganization by Hi-C after mitosis in highly purified, synchronous mouse erythroid cell populations. We observed rapid establishment of A/B compartments, followed by their gradual intensification and expansion. Contact domains form from the ‘bottom up’—smaller subTADs are formed initially, followed by convergence into multi-domain TAD structures. CTCF is partially retained on mitotic chromosomes and immediately resumes full binding in ana/telophase. By contrast, cohesin is completely evicted from mitotic chromosomes and regains focal binding at a slower rate. The formation of CTCF/cohesin co-anchored structural loops follows the kinetics of cohesin positioning. Stripe-shaped contact patterns—anchored by CTCF—grow in length, which is consistent with a loop-extrusion process after mitosis. Interactions between cis-regulatory elements can form rapidly, with rates exceeding those of CTCF/cohesin-anchored contacts. Notably, we identified a group of rapidly emerging transient contacts between cis-regulatory elements in ana/telophase that are dissolved upon G1 entry, co-incident with the establishment of inner boundaries or nearby interfering chromatin loops. We also describe the relationship between transcription reactivation and architectural features. Our findings indicate that distinct but mutually influential forces drive post-mitotic chromatin reconfiguration.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All figures include publicly available data. The Hi-C, Capture-C and ChIP–seq data generated and analysed in this study are deposited in the Gene Expression Omnibus repository under accession number GSE129997 for public access. Additional external ChIP–seq data previously reported are available at: H3K27ac (GSE61349)25, H3K4me1 (GSM946535)26, H3K4me3 (GSM946533)26, H3K36me3 (GSM946529)26 and H3K9me3 (GSM946542)26. CTCF peak files from 13 different tissues are available through the ENCODE project (https://www.encodeproject.org/) with accession numbers ENCFF001LFU, ENCFF001LHE, ENCFF001LHY, ENCFF001LJL, ENCFF001LKO, ENCFF001LMN, ENCFF001LNK, ENCFF001LOR, ENCFF001LPI, ENCFF001LQB, ENCFF001LQS, ENCFF001LSE and ENCFF001LSW.
Code availability
Code used in this study is available upon request from the corresponding authors.
References
Naumova, N. et al. Organization of the mitotic chromosome. Science 342, 948–953 (2013).
Gibcus, J. H. et al. A pathway for mitotic chromosome formation. Science 359, eaao6135 (2018).
Nagano, T. et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature 547, 61–67 (2017).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Weiss, M. J., Yu, C. & Orkin, S. H. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17, 1642–1651 (1997).
Dileep, V. et al. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 25, 1104–1113 (2015).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Norton, H. K. et al. Detecting hierarchical genome folding with network modularity. Nat. Methods 15, 119–122 (2018).
Li, B. et al. A comprehensive mouse transcriptomic bodymap across 17 tissues by RNA-seq. Sci. Rep. 7, 4200 (2017).
Yu, W., He, B. & Tan, K. Identifying topologically associating domains and subdomains by Gaussian Mixture model And Proportion test. Nat. Commun. 8, 535 (2017).
Sanborn, A. L. et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl Acad. Sci. USA 112, E6456–E6465 (2015).
Fudenberg, G. et al. Formation of chromosomal domains by loop extrusion. Cell Reports 15, 2038–2049 (2016).
Oomen, M. E., Hansen, A. S., Liu, Y., Darzacq, X. & Dekker, J. CTCF sites display cell cycle-dependent dynamics in factor binding and nucleosome positioning. Genome Res. 29, 236–249 (2019).
Owens, N. et al. CTCF confers local nucleosome resiliency after DNA replication and during mitosis. eLife 8, e47898 (2019).
Cai, Y. et al. Experimental and computational framework for a dynamic protein atlas of human cell division. Nature 561, 411–415 (2018).
Hughes, J. R. et al. Analysis of hundreds of cis-regulatory landscapes at high resolution in a single, high-throughput experiment. Nat. Genet. 46, 205–212 (2014).
Vian, L. et al. The energetics and physiological impact of cohesin extrusion. Cell 173, 1165–1178.e20 (2018).
Rowley, M. J. et al. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852.e7 (2017).
Hsiung, C. C.-S. et al. A hyperactive transcriptional state marks genome reactivation at the mitosis-G1 transition. Genes Dev. 30, 1423–1439 (2016).
Behera, V. et al. Interrogating histone acetylation and BRD4 as mitotic bookmarks of transcription. Cell Rep. 27, 400–415.e5 (2019).
Yusufzai, T. M., Tagami, H., Nakatani, Y. & Felsenfeld, G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13, 291–298 (2004).
Weintraub, A. S. et al. YY1 is a structural regulator of enhancer–promoter loops. Cell 171, 1573–1588 (2017).
Beagan, J. A. et al. YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res. 27, 1139–1152 (2017).
Schwarzer, W. et al. Two independent modes of chromatin organization revealed by cohesin removal. Nature 551, 51–56 (2017).
Hsu, S. C. et al. The BET protein BRD2 cooperates with CTCF to enforce transcriptional and architectural boundaries. Mol. Cell 66, 102–116 (2017).
Dogan, N. et al. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than histone modifications or chromatin accessibility. Epigenetics Chromatin 8, 16 (2015).
Wu, W. et al. Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 24, 1945–1962 (2014).
Acknowledgements
We thank members of the Blobel and Phillips-Cremins laboratories for discussions, E. Apostolou and J. Dekker for discussing data before publication, L. Mirny for insights, the CHOP flow core facility staff and A. Stout for expert technical support. This work was supported by grants R37DK058044 to G.A.B., R24DK106766 to G.A.B. and R.C.H., U01HL129998A to J.E.P.-C. and G.A.B., The New York Stem Cell Foundation to J.E.P.-C., the NIH Director’s New Innovator Award from the National Institute of Mental Health (1DP2MH11024701; J.E.P.-C), and a generous gift from the DiGaetano family to G.A.B. J.E.P.-C. is a New York Stem Cell Foundation (NYSCF) Robertson Investigator. We acknowledge support from the Spatial and Functional Genomics program at The Children’s Hospital of Philadelphia.
Author information
Authors and Affiliations
Contributions
H.Z., J.E.P.-C. and G.A.B. conceived the study and designed experiments. H.Z. performed experiments with help from P.H., H.W., C.A.K., B.G. and R.C.H. D.J.E. performed initial Hi-C data pre-processing and domain calling. H.Z. performed A/B compartment and ChIP–seq related analysis with help from Y.L. T.G.G. performed loop calling, classification and clustering, and aggregated peak analysis and aggregated domain analysis. K.R.T. performed stripe calling related analysis. D.Z. contributed to Capture-C related analysis. H.Z., J.E.P.-C. and G.A.B. wrote the paper with input from all authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks David Gilbert and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Models, experimental workflow and data quality control.
a, From top to bottom: schematic illustration of the early emergence, gradual intensification and expansion of A/B compartments (checkerboards) from prometaphase to late G1 phase, coupled with schematics of chromatin organization; subTADs (small triangles) emerge first after mitotic exit, followed by convergence into a TAD (big triangle); formation of a structural loop coincides with the positioning of cohesin, but not CTCF after mitosis; the gradual extrusion of cohesin complex along DNA fibre from one anchor point with CTCF, reflected as enrichment of interactions between the anchor and a continuum of DNA loci on the contact map; fast formation of E/P loops after mitosis; the interplay between transient E/P loops and boundaries or structural loops. b, The experimental workflow. Representative flow cytometry plots showing the nocodazole arrest–release strategy based on pMPM2 (prometaphase), mCherry–MD signal, and DNA content (DAPI) staining. Similar observations were made in more than 5 independent experiments. c, Representative images showing DAPI and lamin B1 staining of FACS-purified cells across all stages of the cell cycle. Similar observations were made in 2 independent experiments. The mitotic index of prometaphase cells after FACS purification is on average greater than 98%. Yellow and white arrowheads indicate anaphase and telophase cells, respectively. Scale bar, 10 μm. d, Hexbin plots showing the high correlation of Hi-C raw read counts between two biological replicates across all stages of the cell cycle. Bin size, 250 kb. e, Heat map showing the Pearson correlation among all Hi-C samples, based on the eigenvector 1 of 250 kb bins. f, Heat map showing the Pearson correlation among all Hi-C samples based on raw read counts. Bin size, 250 kb. g–i, Heat maps showing Pearson correlation of CTCF (g), Rad21 (h) and Pol II (i) ChIP–seq data among all samples. Note the overall high replicate concordance. Low correlation coefficients among replicates were only observed in samples with low signal-to-noise ratios—for example, in prometaphase.
Extended Data Fig. 2 Compartment strengthening and expansion from ana/telophase throughout late G1.
a, Saddle plots showing the progressive gain of compartment strength over time in two biological replicates. b, Schematic showing the calculation of compartment strength. c, Line graphs showing the progressive increase of compartment strength of each individual chromosome (represented by dots) in two biological replicates. d, Heat map showing the genome-wide Spearman correlation coefficients between eigenvector 1 values and asynchronous-cell-derived ChIP–seq signals for the indicated histone marks. e, Plots of chromosome-averaged distance-dependent contact frequency (P(s)) at all stages of the cell cycle. f, P(s) plots of each individual chromosome (two biological replicates). g, A schematic illustrating how compartmentalization levels (R(s)) were calculated at different distance scales (for example, 1 Mb or 100 Mb). Each dotted line indicates a series of 250-kb bin–bin pairs that are separated by a given genomic distance s (the distance from the diagonal to the dotted line). For all bin–bin pairs separated by distance of s, a Spearman correlation coefficient R(s) was generated between observed/expected and the product of two eigenvector 1 values (PC1 (bin1) × PC1 (bin2)). R(s) is expected to be high in well-compartmentalized regions (left) and low at large distance scales with no compartments (right). h, Replicate-averaged R(s) of each individual chromosome across all stages of the cell cycle when s is equal to 10, 50 and 125 Mb (only eight chromosomes were computed at the 125-Mb scale). i, Line graph showing the level of compartmentalization of chr1 against genomic distance at each stage of the cell cycle.
Extended Data Fig. 3 Domain detection and residual ‘domain-like’ structures in prometaphase.
a, b, Meta-region plots and density heat maps of insulation scores (a) and directionality index (b) centred around domain boundaries initially detected at each stage of the cell cycle. c, Scatter plots showing Pearson correlations of insulation scores at domain boundaries between two biological replicates. d, Aggregated domain analysis (ADA) of domains initially detected at each stage of the cell cycle. e, Box plots showing ADA scores over time for domains initially detected at prometaphase (n = 1,360), ana/telophase (n = 2,260), early G1 (n = 2,875) and mid G1 (n = 1,112). For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Mann–Whitney U-test. The dotted line indicates the average ADA score of initial domain detection. f, Hi-C contact maps of two representative regions (chr8: 113 Mb–114 Mb and chr9: 72 Mb–73 Mb) showing residual domain- and boundary-like structures (yellow lines) in prometaphase in merged and individual biological replicates. Bin size, 10 kb. g, Simulated featureless, per cent ‘G1 contaminated’, and early G1 contact maps of the same regions as in f. Bin size, 10 kb. h, Meta-region plots showing the insulation scores of prometaphase, simulated featureless, ‘G1-contaminated’ and early G1 samples, centred around prometaphase boundaries in chr8 and chr9. i, Meta-region plots showing indicated histone modification profiles centred around boundaries newly detected at each stage of the cell cycle. j, Bar graphs showing the enrichment of transcription start sites (overall, housekeeping and tissue-specific9) within ±20 kb of boundaries newly detected at each stage of the cell cycle.
Extended Data Fig. 4 Dynamics of TAD and subTAD after mitosis.
a, Schematic of possible models of hierarchical domain formation: bottom-up (merge), top-down (split) and concomitant. b, Bar graphs showing the fraction of TADs that display either type of behaviour after detection. c, Bar graphs showing the fraction of subTADs that display each of the four potential behaviours after detection: merge, split, merge and split, and static. d, Bottom, schematic showing partitioning of boundaries into TAD and subTAD boundaries. Top, Hi-C contact maps showing the change in insulation of representative TAD and subTAD boundaries from ana/telophase to late G1. SubTAD and TAD boundaries are indicated by green and blue arrows, respectively. Bin size, 10 kb. e, Bin plots showing the change in insulation score over time of TAD boundaries (top) and subTAD boundaries (bottom) that are detected at prometaphase in merged replicates and in two biological replicates. f, Box plots showing sizes of domains initially detected at prometaphase (n = 2,494), ana/telophase (n = 1,699), early G1 (n = 1,357) and mid G1 (n = 682) by rGMAP. For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Mann–Whitney U-test. g, Pie charts of the cell cycle distribution of subTADs and TADs that contain at least 1 subTAD based on their time of emergence (called by rGMAP). The P value was calculated using a two-sided Fisher’s exact test (prometaphase + ana/telophase compared with early G1 + mid G1). h, Bar graphs showing the fraction of subTADs detected by rGMAP that display each of the four potential behaviours after detection: merge, split, merge and split, and static. i, Bin plots showing the change in insulation score of TAD boundaries (left) and subTAD boundaries (right) that are detected by rGMAP at prometaphase. j, Box plots showing the sizes of domains initially detected at prometaphase (n = 1,105), ana/telophase (n = 1,124), early G1 (n = 2,385) and mid G1 (n = 520) by DI+sweep (directionality index + window size adjustment). For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated by two-sided Mann–Whitney U-test. k–m, Similar to g–i, showing analyses based on domains called by DI+sweep.
Extended Data Fig. 5 CTCF and cohesin chromatin occupancy in mitosis and G1 entry.
a, A density heat map of CTCF ChIP–seq data of each biological replicate of asynchronous and prometaphase samples, centred around IM- and IO-CTCF binding sites. b, A density heat map of Rad21 ChIP–seq data of both biological replicates of asynchronous and prometaphase samples centred around all Rad21 peaks. c, Genome browser tracks showing CTCF and Rad21 ChIP–seq signals of asynchronous and prometaphase samples at indicated regions. n = 2–3 biological replicates. d, ChIP–qPCR data of CTCF and Rad21 in asynchronous (n = 3, 6 biological replicates for CTCF and Rad21, respectively) and prometaphase samples (n = 4, 3 biological replicates for CTCF and Rad21, respectively). Data are mean ± s.e.m. e, Motif enrichment analysis of IM- and IO-CTCF binding sites with indicated E values as determined by MEME-ChIP. f, Top, donut charts showing the genome-wide distribution of IM- and IO-CTCF binding sites. Middle, bar graphs showing the percentage of IM- or IO-CTCF-binding sites that are found in indicated numbers of tissues. Bottom, donut pie chart showing the fraction of IM- and IO-CTCF binding sites that are co-occupied by Rad21. g, Density heat maps and meta-region plots of CTCF and Rad21 ChIP–seq data across all time points centred around CTCF-specific and CTCF/Rad21 co-occupied binding sites. h, Bin plots showing ChIP–seq signals of CTCF and Rad21 peaks for each stage of the cell cycle (y axis) against late G1 (x axis). i, ChIP–qPCR of CTCF and Rad21 at indicated binding sites over time. n = 2 biological replicates for 0 and 25 min, and n = 3 biological replicates for 120 and 240 min after nocodazole release. Data are mean ± s.e.m. j, Schematic showing mCherry-tagging of endogenous CTCF and SMC3. k, Representative images (from at least 10 dividing cells) illustrating the behaviour of mCherry-tagged CTCF and SMC3 during mitosis–early G1 phase progression. Similar observations were made in 2 independent experiments. Yellow dotted circles demarcate cell nuclei after mitosis. Scale bar, 5 μm. l, Average recovery curve of mCherry-tagged CTCF and SMC3 that co-localize with H2B–YFP. Cells (11 mother cells/22 daughter cells and 10 mother cells/18 daughter cells) were analysed for CTCF and SMC3, respectively. P values were calculated using a two-sided Student’s t-test. Data are mean ± s.e.m. P values were omitted at time points with fewer than 5 cells.
Extended Data Fig. 6 Loop statistics and k-means clustering on structural loops.
a, Bar graph showing the number of loop calls at each stage of the cell cycle. b, Aggregated peak analysis (APA) of loops initially detected at each stage of the cell cycle. Bin size, 10 kb. Numbers indicate average loop strength: ln(obs/exp). c, Scatter plots showing the Pearson correlation of loop strength (read counts) between two biological replicates. d, Hi-C contact maps showing representative regions that contain cluster 1 (chr1: 172.8 Mb–173 Mb), 2 (chr1: 90.2 Mb–90.8 Mb) and 3 (chr2: 47.5 Mb–49 Mb) structural loops in merged and both biological replicates. Bin size, 10 kb. Loop signal enrichment is indicated by black arrows. Contact maps are coupled with genome browser tracks showing CTCF and cohesin occupancy across all stages of the cell cycle. Chevron arrows mark orientations of CTCF sites at loop anchors. e, APA of cluster 1, 2 and 3 structural loops across all stages of the cell cycle. Each heat map is coupled with four meta-region plots corresponding to CTCF and Rad21 ChIP–seq signals centred around either upstream or downstream loop anchors. Bin size, 10 kb. Numbers indicate average loop strength: ln(obs/exp). f, Left and right, schematics showing how correlations are computed between CTCF or Rad21 and loop strength over time. Middle, box plot showing the Pearson correlation coefficients between CTCF or Rad21 ChIP–seq peak strength at upstream or downstream anchors and structural loop strength over time (n = 4,712). For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Wilcoxon signed-rank test. g, Box plot showing sizes of structural loops initially detected at ana/telophase (n = 90), early G1 (n = 2,233), mid G1 (n = 1,595) and late G1 (n = 793). For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Mann–Whitney U-test. h, Average recovery curves of structural loops (n = 4,241) and E/P loops with 0 (n = 678) or 1 (n = 1,338) anchor co-occupied by CTCF/cohesin. The 10% of loops with the smallest increment from prometaphase to late G1 were filtered out from the analysis. Data are mean ± 99% confidence interval. **** and ####, P < 2.2 × 10−16 (structural loops compared with E/P loops with 0 or 1 anchor co-occupied by CTCF/cohesin, respectively). Two-sided Mann–Whitney U-test. i, Left, average recovery curves of randomly sampled and size-matched structural loops and CTCF/cohesin independent E/P loops (n = 2,869 for both groups). The 10% of loops with the smallest increment from prometaphase to late G1 were filtered out from the analysis. Data are mean ± 99% confidence interval. P values were calculated using a two-sided Mann–Whitney U-test. Right, box plot showing the comparable size distribution of these two randomly sampled groups (n = 2,869 for both). For both box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. j, Bar graphs depicting the composition of loops newly called at each stage of the cell cycle.
Extended Data Fig. 7 Reformation of chromatin stripes after mitosis.
a, Pie chart showing the fraction of stripes with inwardly oriented CTCF at stripe anchors. b, Hi-C contact maps of two representative regions (chr2: 12.75 Mb–14.75 Mb and chr1: 130.5 Mb–132.5 Mb) that contain stripes with inwardly oriented CTCF. Bin size, 10 kb. Contact maps are coupled with genome browser tracks of CTCF and Rad21 across all stages of the cell cycle and tracks of asynchronous H3K4me3, H3K4me1 and H3K27ac and annotation of cis-regulatory elements. Chevron arrows mark positions and orientations of CTCF peaks at stripe and loop anchors. Lengthening of stripes is indicated by black arrows. Stripe anchors are indicated by purple arrows. Loops along the stripe axis and at the far end of stripes are indicated by blue circles. c, similar to b, Hi-C contact maps showing a representative stripe (chr10: 118.2 Mb–118.8 Mb) that does not have inwardly oriented CTCF at the stripe anchor. d, Left, aggregated Hi-C contact maps that compile all stripes with inwardly oriented CTCF to show their overall dynamic growth after mitosis. Right, box plots showing the lengths of these stripes at ana/telophase (n = 235), early G1 (n = 1,472), mid G1 (n = 1,477) and late G1 (n = 1,473). For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Mann–Whitney U-test. e, Similar to d, showing stripes without inwardly oriented CTCF. n = 72, 281, 277, 272 for ana/telophase, early G1, mid G1 and late G1, respectively. f, H3K27ac ChIP–seq profile from asynchronous G1E-ER4 cells is plotted −200 kb to 2 Mb around the horizontal stripe anchors and −2 Mb to 200 kb around the vertical stripe anchors. Anchor position is indicated by purple arrows.
Extended Data Fig. 8 Supplementary E/P loop analyses.
a, APA of the three clusters of E/P loops on merged and two biological replicates. Bin size, 10 kb. Numbers indicate average loop strength: ln(obs/exp). b, Hi-C contact maps showing an additional example of cluster 1 E/P loop (chr1: 43.45 Mb–43.65 Mb, green arrow). Bin size, 10 kb. Colour bar denotes q-normed reads. Contact maps are coupled with genome browser tracks of CTCF and cohesin across all time points as well as asynchronous H3K4me3, H3K4me1 and H3K27ac and annotations of cis-regulatory elements. c, Similar to b, showing two examples of manually identified transient E/P contacts (Pde12 locus and Morc3 locus, indicated by red arrow). Boundaries or structural loop anchors that potentially interfere with these E/P contacts are indicated by black and blue arrows, respectively. Contact maps are coupled with tracks of Capture-C interaction profiles. Probes (anchor symbol) are located at promoters of Pde12 and Morc3 genes. d, Hi-C contact maps showing the Pde12 locus on two biological replicates. Bin size, 10 kb. e, Quantification of the Capture-C read density of the red regions in c. n = 3 biological replicates. Data are mean ± s.e.m. P values were calculated from two-sided Student’s t-test. f, Similar to d, Hi-C contact maps showing the cluster3 E/P loop (red arrows) at Commd3 locus in two biological replicates. Potential interfering loop is indicated by blue arrows. g, Insulation score profiles centred around the boundaries and interfering structural loop anchors that solely reside within cluster 1, 2 or 3 E/P loops. h, Sanger sequencing profiles showing deletion of the CTCF core motif at the upstream anchor of the structural loop (blue arrows in f) that potentially interfere with the cluster3 E/P loop at the Commd3 locus (red arrows in f). i, ChIP–qPCR showing the abrogation of CTCF and Rad21 binding at the edited site in f. n = 3 biological replicates. Data are mean ± s.e.m. P values were calculated by two-sided Student’s t-test. j, Schematic showing potential behaviour of cluster 3 E/P loops before and after deletion of the interfering structural loop anchor. k, Capture-C interaction profiles between Commd3 promoter and downstream cis-regulatory element (red bars) on wild-type and interfering anchor-deleted mutant cells over time. The location of the capture probe is indicated by the anchor symbol. The deleted CTCF site is indicated by green triangles. Formation of the transient loop is indicated by red arches. l, Quantification showing read density of the red regions in k. n = 3 and 2 biological replicates for wild-type and mutant cells, respectively. Data are mean ± s.e.m. P values were calculated by two-sided Student’s t-test. m, Box plots showing ChIP–seq signals of indicated histone modifications at anchors that solely participate in cluster 1, 2 or 3 (transient) E/P loops (n = 2,612, 1,338 and 413 respectively). For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Mann–Whitney U-test.
Extended Data Fig. 9 Relationship between post-mitotic structural organization and gene reactivation.
a, Meta-region analysis of Pol II occupancy of active genes across all stages of the cell cycle. TSS, transcription start site; TES, transcription end site. b, Bin plots showing the positive correlation between Pol II ChIP–seq signal strength and eigenvector 1 (asynchronous G1E-ER4 cells27, 25-kb binned) genome-wide. c, Left, schematic showing genes that are within early or late domains. Right, average Pol II occupancy of genes that reside in prometaphase (n = 2,274 genes) ana/telophase (n = 2,114 genes), early G1 (n = 1,159 genes) and mid G1 (n = 303 genes) emerging domains. Data are mean ± 99% confidence interval. d, Heat map showing gene-body Pol II occupancy across all stages of the cell cycle. Genes are ranked by their PC1 values (‘spikiness’). e, Genome browser tracks showing representative examples of early spiking (Kpna2) and gradually activating (Nedd4) genes. f, Quantification of gene-body Pol II occupancy in e. n = 2 biological replicates for 0 h, and n = 3 biological replicates for other time points. Data are mean ± s.e.m. g, Schematic showing the stratification of genes on the basis of their involvement in E/P loops. h, Table showing the number of genes that are solely involved in clusters of E/P loops. i, Average gene-body Pol II occupancy of the genes in h over time. Sample sizes are shown in h. Data are mean ± s.e.m. j, Box plots showing the spikiness (PC1) of genes in h. Sample sizes are shown in h. For all box plots, centre lines denote medians; box limits denote 25th–75th percentile; whiskers denote 5th–95th percentile. P values were calculated using a two-sided Mann–Whitney U-test.
Supplementary information
Supplementary Methods
This file contains Supplementary Methods and references.
Supplementary Table 1
Overall domain calls and domain calls at each cell cycle stage.
Supplementary Table 2
CTCF & Rad21 union peak list.
Supplementary Table 3
Overall loop calls.
Supplementary Table 4
Active gene list.
Supplementary Table 5
Oligo sequences.
Supplementary Table 6
Hi-C data processing statistics.
Supplementary Table 7
ChIP-seq data processing statistics.
Rights and permissions
About this article
Cite this article
Zhang, H., Emerson, D.J., Gilgenast, T.G. et al. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576, 158–162 (2019). https://doi.org/10.1038/s41586-019-1778-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1778-y
This article is cited by
-
3D genomics and its applications in precision medicine
Cellular & Molecular Biology Letters (2023)
-
Live-cell imaging of chromatin contacts opens a new window into chromatin dynamics
Epigenetics & Chromatin (2023)
-
Sox11 is enriched in myogenic progenitors but dispensable for development and regeneration of the skeletal muscle
Skeletal Muscle (2023)
-
SARS-CoV-2 restructures host chromatin architecture
Nature Microbiology (2023)
-
The spatial organization of transcriptional control
Nature Reviews Genetics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.