Abstract
Bacteria have evolved sophisticated mechanisms to inhibit the growth of competitors1. One such mechanism involves type VI secretion systems, which bacteria can use to inject antibacterial toxins directly into neighbouring cells. Many of these toxins target the integrity of the cell envelope, but the full range of growth inhibitory mechanisms remains unknown2. Here we identify a type VI secretion effector, Tas1, in the opportunistic pathogen Pseudomonas aeruginosa. The crystal structure of Tas1 shows that it is similar to enzymes that synthesize (p)ppGpp, a broadly conserved signalling molecule in bacteria that modulates cell growth rate, particularly in response to nutritional stress3. However, Tas1 does not synthesize (p)ppGpp; instead, it pyrophosphorylates adenosine nucleotides to produce (p)ppApp at rates of nearly 180,000 molecules per minute. Consequently, the delivery of Tas1 into competitor cells drives rapid accumulation of (p)ppApp, depletion of ATP, and widespread dysregulation of essential metabolic pathways, thereby resulting in target cell death. Our findings reveal a previously undescribed mechanism for interbacterial antagonism and demonstrate a physiological role for the metabolite (p)ppApp in bacteria.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data supporting the findings of this study are available within the manuscript and associated Supplementary Information. X-ray crystallographic coordinates and structure factor files are available from the PDB with the following accession numbers: Tas1tox–Tis1 (6OX6) and PurFEC-ppApp (6OTT). Maximum likelihood estimates of P. aeruginosa strain relationships used for tree construction are provided in Newick format in Supplementary Dataset 1. Relative concentrations of metabolites from metabolomics are reported in Supplementary Dataset 2. Source gel images are available in Supplementary Fig. 1.
References
Granato, E. T., Meiller-Legrand, T. A. & Foster, K. R. The evolution and ecology of bacterial warfare. Curr. Biol. 29, R521–R537 (2019).
Russell, A. B., Peterson, S. B. & Mougous, J. D. Type VI secretion system effectors: poisons with a purpose. Nat. Rev. Microbiol. 12, 137–148 (2014).
Hauryliuk, V., Atkinson, G. C., Murakami, K. S., Tenson, T. & Gerdes, K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 13, 298–309 (2015).
Wexler, A. G. et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–3644 (2016).
Whitney, J. C. et al. Genetically distinct pathways guide effector export through the type VI secretion system. Mol. Microbiol. 92, 529–542 (2014).
Whitney, J. C. et al. An interbacterial NAD(P)+ glycohydrolase toxin requires elongation factor Tu for delivery to target cells. Cell 163, 607–619 (2015).
Quentin, D. et al. Mechanism of loading and translocation of type VI secretion system effector Tse6. Nat. Microbiol. 3, 1142–1152 (2018).
Johnson, L. S., Eddy, S. R. & Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11, 431 (2010).
Atkinson, G. C., Tenson, T. & Hauryliuk, V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6, e23479 (2011).
Potrykus, K., Murphy, H., Philippe, N. & Cashel, M. ppGpp is the major source of growth rate control in E. coli. Environ. Microbiol. 13, 563–575 (2011).
Wang, B. et al. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150 (2019).
Gaca, A. O. et al. From (p)ppGpp to (pp)pGpp: characterization of regulatory effects of pGpp synthesized by the small alarmone synthetase of Enterococcus faecalis. J. Bacteriol. 197, 2908–2919 (2015).
Beljantseva, J. et al. Negative allosteric regulation of Enterococcus faecalis small alarmone synthetase RelQ by single-stranded RNA. Proc. Natl Acad. Sci. USA 114, 3726–3731 (2017).
Sarubbi, E. et al. Characterization of the spoT gene of Escherichia coli. J. Biol. Chem. 264, 15074–15082 (1989).
Bugg, T. D., Braddick, D., Dowson, C. G. & Roper, D. I. Bacterial cell wall assembly: still an attractive antibacterial target. Trends Biotechnol. 29, 167–173 (2011).
Raetz, C. R. Enzymology, genetics, and regulation of membrane phospholipid synthesis in Escherichia coli. Microbiol. Rev. 42, 614–659 (1978).
LaCourse, K. D. et al. Conditional toxicity and synergy drive diversity among antibacterial effectors. Nat. Microbiol. 3, 440–446 (2018).
Rhaese, H. J. & Groscurth, R. Control of development: role of regulatory nucleotides synthesized by membranes of Bacillus subtilis in initiation of sporulation. Proc. Natl Acad. Sci. USA 73, 331–335 (1976).
Rhaese, H. J., Hoch, J. A. & Groscurth, R. Studies on the control of development: isolation of Bacillus subtilis mutants blocked early in sporulation and defective in synthesis of highly phosphorylated nucleotides. Proc. Natl Acad. Sci. USA 74, 1125–1129 (1977).
Goodman, A. L. et al. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754 (2004).
Marden, J. N. et al. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 110, 15055–15060 (2013).
Steinchen, W. et al. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc. Natl Acad. Sci. USA 112, 13348–13353 (2015).
McGinness, K. E., Baker, T. A. & Sauer, R. T. Engineering controllable protein degradation. Mol. Cell 22, 701–707 (2006).
Stover, C. K. et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406, 959–964 (2000).
Lee, D. G. et al. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 7, R90 (2006).
Rietsch, A., Vallet-Gely, I., Dove, S. L. & Mekalanos, J. J. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 102, 8006–8011 (2005).
Hmelo, L. R. et al. Precision-engineering the Pseudomonas aeruginosa genome with two-step allelic exchange. Nat. Protocols 10, 1820–1841 (2015).
Winsor, G. L. et al. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 44, D646–D653 (2016).
Hyatt, D. et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11, 119 (2010).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 (2009).
Treangen, T. J., Ondov, B. D., Koren, S. & Phillippy, A. M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524 (2014).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Hood, R. D. et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 (2010).
Stevens, A. J. et al. Design of a split intein with exceptional protein splicing activity. J. Am. Chem. Soc. 138, 2162–2165 (2016).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D 62, 859–866 (2006).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
Bunkóczi, G. et al. Phaser.MRage: automated molecular replacement. Acta Crystallogr. D 69, 2276–2286 (2013).
Manav, M. C. et al. Structural basis for (p)ppGpp synthesis by the Staphylococcus aureus small alarmone synthetase RelP. J. Biol. Chem. 293, 3254–3264 (2018).
Acknowledgements
We thank A. Raphenya and B. Alcock for assistance with sequence data curation and analyses, C. Chang for assistance with X-ray data collection and processing, and the Whitehead Institute Metabolite Profiling Core Facility for measuring metabolite levels. S.A. and B.W. were supported by an Ontario Graduate Scholarship and a fellowship from the Jane Coffin Childs Memorial Fund, respectively. A.G.M. holds a Cisco Research Chair in Bioinformatics and M.T.L is an Investigator of the Howard Hughes Medical Institute. Results shown in this report are derived from work performed by the Structural Biology Center (SBC) and the Northeastern Collaborative Access Team (NECAT) at the Advanced Photon Source, Argonne National Laboratory. SBC is funded by NIAID (HHSN272201200026C) and HHS (HHSN272201700060C) and NECAT is funded by NIH grants P30 GM124165 and S10OD021527. SBC-CAT and NECAT are operated by UChicago Argonne, LLC, for the US DOE under contract number DE-AC02-06CH11357. This work was supported by grants from the Canadian Foundation for Innovation (34531 to A.G.M.), NIH (R01-GM082899 to M.T.L.) and CIHR (PJT-156129 to J.C.W.), and by seed funding from the David Braley Centre for Antibiotic Discovery (to J.C.W.).
Author information
Authors and Affiliations
Contributions
Experiments were conceived and designed by S.A., B.W., M.T.L. and J.C.W. Cloning, bacterial competition assays, protein purification, biochemical experiments and protein crystallization were carried out by S.A. and B.W. X-ray data collection and analyses were performed by P.J.S. and R.A.G. Bioinformatics analyses for Extended Data Fig. 1a were performed by H.-K.R.T. and A.G.M. Assistance with cloning, purification and crystallization of Tas1–Tis1 complex was provided by M.D.W. Figure design, manuscript writing and editing were done by S.A., B.W., M.T.L. and J.C.W. The project was supervised by M.T.L. and J.C.W. Funding was provided by A.S., R.A.G., A.G.M., M.T.L. and J.C.W.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Urs Jenal, Justin Nodwell and Jue Wang for their contribution to the peer review of this work.
Extended data figures and tables
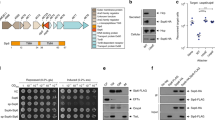
Extended Data Fig. 1 Homologues of PA14_01140 and Tse6 are enriched in P. aeruginosa PA14- and PAO1-related strains, respectively.
a, Phylogenetic distribution of PA14_01140 (pink) and tse6 (blue) within 326 P. aeruginosa genomes based on whole-genome single-nucleotide polymorphism (SNP) maximum likelihood analysis. Circles denote individual P. aeruginosa strains. Each clade is labelled according to its representative member. The miniaturized tree depicts true branch distance between each clade. The full tree in Newick format, including bootstrap values, is provided as Supplementary Dataset 1. b, Proteins containing a domain homologous to the C terminus of PA14_01140 are found in several species of Proteobacteria. Homologues were identified using the HMMER webserver and candidate T6SS effectors were selected on the basis of the presence of predicted N-terminal domains known to facilitate export by the T6SS. The UniProt accession number for each identified protein is indicated.
Extended Data Fig. 2 Characterization of the PA14_01140–PA14_01130–tsi6 gene cluster.
a, Expression of the conserved H1-T6SS effector Tse1 and the secreted H1-T6SS subunit Hcp1 are similar between P. aeruginosa PAO1 ΔretS and P. aeruginosa PA14 ΔrsmAΔrsmF by western blot analysis. A non-specific band that reacts with the anti-Tse1 antiserum was used as a loading control. b, Tsi6PA14 is not protective against Tse6-mediated intoxication. Viability of E. coli cells grown on solid medium harbouring inducible plasmids expressing Tse6tox, Tse6tox + Tsi6PAO1, Tse6tox + Tsi6PA14, or an empty vector control. c, Mutational inactivation of PA14_01140 does not abrogate secretion of Hcp1. Western blot analysis of Hcp1 in the cell and supernatant (sup) fractions of the indicated P. aeruginosa PA14 strains. d, Delivery of PA14_01140 into recipient cells requires the H1-T6SS exported protein VgrG1 and the Tse6-specific chaperone EagT6. Intraspecific growth competition assay between indicated PA14 donor and recipient strains. The parental strain genotype is ΔrsmAΔrsmF. Mean ± s.d. for n = 3 biological replicates; two-tailed, unpaired t-test. e, Mutational inactivation of eagT6, vgrG1, vgrG2 and vgrG4 does not abrogate H1-T6SS function. Western blot analysis of Hcp1 in the cell and supernatant fractions of the indicated P. aeruginosa PA14 strains. a–c, e, Data are representative of three independent experiments.
Extended Data Fig. 3 PA14_01140tox possesses remote homology to characterized (p)ppGpp synthetases.
ClustalW alignment of PA14_01140tox, the RSH domains of E. coli RelA and Streptococcus equisimilis Rel, and the small alarmone synthetases RelQ and RelP from B. subtilis and S. aureus, respectively. Dashed boxes represent regions of high sequence homology. The catalytic glutamic acid is indicated by a red triangle.
Extended Data Fig. 4 The C-terminal domain of PA14_01140 (PA14_01140tox) is toxic when expressed in E. coli.
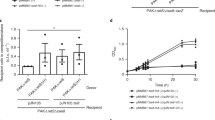
a, PA14_01130 but not Tsi6PA14 inhibits PA14_01140tox-mediated toxicity. Viability of E. coli cells grown on solid medium harbouring inducible plasmids expressing PA14_01140tox, PA14_01140tox + PA14_01130, PA14_01140tox + Tsi6PA14, or an empty vector control. b, c, PA14_01140tox is toxic to E. coli, even when expressed at approximately three copies per cell. b, Western blot analysis of pull-downs from E. coli expressing His6–PA14_01140tox-VSV-G in the presence of the indicated concentrations of aTC inducer (see Methods). c, Viability of E. coli cells expressing His6–PA14_01140tox-VSV-G in the presence of the indicated aTC concentrations for 15 min. d, Amino acid residues in PA14_01140tox that structurally align with known pyrophosphate donor ATP-interacting residues in RelQ are required for PA14_01140tox-mediated toxicity. Viability of E. coli cells grown on solid medium harbouring inducible plasmids expressing PA14_01140tox, each of the indicated PA14_01140tox point mutants or an empty vector control. Lysine 326 is a residue located within the PA14_01140tox active site that is not predicted to interact with the pyrophosphate donor ATP. e, Glutamate 382 is required for PA14_01140-based intoxication of susceptible recipient cells. Outcome of intraspecific growth competitions between the indicated PA14 donor strains and a ΔPA14_01130-1140 recipient. The parental PA14 strain genotype is ΔrsmAΔrsmF. The competitive index is normalized to starting donor/recipient ratios. Mean ± s.d. for n = 3 biological replicates; two-tailed, unpaired t-test. a–e, Data are representative of two independent experiments.
Extended Data Fig. 5 Interaction with PA14_01130 distorts the predicted nucleotide acceptor site of PA14_01140tox.
Structural alignment between PA14_01140tox–PA14_01130 complex and the (p)ppGpp synthetase RelP bound to the non-hydrolysable ATP analogue AMPCPP and a GTP acceptor nucleotide (PDB code 6EWZ)40. Two C-terminal α-helices of PA14_01140tox that align with the GTP binding site of RelP are rotated approximately 30° as a consequence of their interaction with PA14_01130 (curved black arrow).
Extended Data Fig. 6 Tas1 pyrophosphorylates the 3′ hydroxyl group of adenosine nucleotides.
a, 1H (top) and 31P (bottom) NMR spectra of pApp. See Supplementary Table 2 for assignments. b, Negative mode electrospray mass spectra for pApp (top), ppApp (middle) and pppApp (bottom). Assignment of major peaks is shown below the spectra. c, Anion-exchange traces of Tas1tox-catalysed reactions with dATP or GTP as pyrophosphate donors. Arrowheads indicate 3′ pyrophosphorylation products. a–c, Data are representative of two independent experiments.
Extended Data Fig. 7 Purified Tas1tox can use pppApp as a pyrophosphate donor to pyrophosphorylate AMP and form pApp.
a, Anion-exchange traces of pppApp and AMP after incubation with the indicated concentrations of Tas1tox for 30 min at room temperature. A control lacking Tas1tox is shown for comparison. Chromatogram is representative of two independent experiments. b, Mechanism of quantitative conversion of ATP to pApp. Only heteroatoms that participate in the reaction mechanism of pApp formation are shown.
Extended Data Fig. 8 Tas1tox overexpression in E. coli leads to accumulation of (p)(p)pApp and a reduction in cellular 5′ adenosine nucleotides.
a, Anion-exchange chromatography traces of metabolites extracted from E. coli cells overexpressing Tas1tox (left) or Tas1tox(E382A) (right) at the indicated time points. A trace generated from a mixture of standards containing an equimolar amount of AMP (1), ADP (2), ATP (3), pApp (4), ppApp (5) and pppApp (6) using the same gradient is shown for comparison. Peaks of adenosine 5′-nucleotides (AMP, ADP and ATP) and (p)(p)pApp are indicated by blue and orange arrowheads, respectively. Traces are representative of three independent experiments. b, Quantification of adenosine 5′-nucleotide and (p)(p)pApp levels in the E. coli strains from a as a function of time after induction. Mean ± s.d. for metabolites extracted from n = 3 separate cultures. Asterisks indicate metabolites below the detection limit.
Extended Data Fig. 9 The pmf-uncoupling ionophore CCCP but not the ppGpp-hydrolase domain of SpoT reduces the toxicity of Tas1tox.
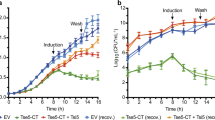
a, Tis1-depleted cells exhibited a reduction in viability over time. CFU plating of P. aeruginosa PA14 ΔretSΔsspB Tis1-D4 pPSV39::sspB cells at the indicated times after induction of SspB expression. b, Tas1 reduces the viability of susceptible recipient cells during interbacterial competition. CFU plating of the indicated P. aeruginosa PA14 recipient strains after co-culture with a parental donor strain at the indicated times. The parental PA14 strain genotype is ΔrsmAΔrsmF. c, Steady-state growth of E. coli is not substantially affected by the presence of carbonyl cyanide m-chlorophenyl hydrazine (CCCP). Growth curves of E. coli cells harbouring the Tas1tox expression plasmid in LB medium with or without CCCP. Curves for n = 3 cultures are overlaid for each condition. d, Tas1tox toxicity is reduced in the presence of CCCP. Viability of E. coli cells following Tas1tox expression in the presence or absence of CCCP. Cells were plated either before induction or at the indicated times after induction. e, Alkaline pH does not affect the ability of CCCP to reduce Tas1tox-dependent toxicity, indicating that the toxicity of Tas1tox is likely to arise from the generation of excessive membrane electrostatic potential. Cultures were untreated or conditioned to pH 8.0 using 25 mM Tris-HCl buffer immediately before induction. f, Activity of the ppGpp-hydrolase domain of SpoT against either ppGpp or ppApp. Initial velocities were normalized to hydrolase activity in the absence of either nucleotide. Mean ± s.d. for enzymatic activity from n = 4 technical replicates; two-tailed, unpaired t-test. g, Anion-exchange chromatography traces of metabolites extracted from growth competition experiments between the indicated strains conducted on solid medium for 4 h. The parental strain is ΔrsmAΔrsmF. Traces are representative of three independent experiments. a, b, d, e, Plates are representative of three independent experiments.
Extended Data Fig. 10 (p)ppApp binds to and inhibits PurF in a similar manner to ppGpp.
a, Isothermal calorimetry trace (top) and fitted isotherm (bottom) for the titration of 100 µM PurFEC with 1 mM pppApp. Data are representative of two independent experiments. b, Changes to the activity of PurFPA in the presence of indicated concentrations of ppGpp or (p)ppApp. Mean ± s.d. for n = 3 separate reactions. c, 2Fo − Fc difference electron density maps of ppApp (left) and ppGpp (right, PDB code 6CZF) contoured at 0.4σ are shown in blue. Nucleotides are shown as stick models of two overlapping ppApp–Mg2+ (coloured by heteroatom or light blue) or ppGpp–Mg2+ (coloured by heteroatom or yellow) complexes, related by a twofold rotational axis. d, Comparison of ppGpp and ppApp binding configuration within PurFEC. The nucleotide–Mg2+ complexes are modelled at 0.5 occupancy because they lie on a crystallographic twofold rotational axis as shown in c. Relevant hydrogen bonding interactions and their distance in angstroms between PurFEC residues and the purine rings of ppApp (left) or ppGpp (right) are shown with red dashed lines.
Supplementary information
Supplementary Tables
This file contains Supplementary Tables 1-5 and Supplementary Figure 1.
Supplementary Data
Supplementary Dataset 1. Maximum likelihood estimate of P. aeruginosa strain relationships constructed using the RAxML-HPC Blackbox. Attached as a separate .txt file in Newick format.
Supplementary Data
Supplementary Dataset 2. Relative metabolite concentrations of P. aeruginosa cells undergoing Tas1-mediated intoxication (n = 3 for Tis1-depleted and control cultures). Attached as a separate .xls file.
Video 1
Time-lapse series of P .aeruginosa PA14 ΔretSΔsspB Tis1-D4 pPSV39::sspB pre-induction. Cells were spotted on an LB growth pad comprised of 1.5% agarose and 2.5μg/mL propidium iodide and imaged every 10 min for a total of 6 hours (n = 1).
Video 2
Time-lapse series of P .aeruginosa PA14 ΔretSsspB Tis1-D4 pPSV39::sspB 2-hr following induction of Tis1 depletion. Cells were spotted on an LB growth pad comprised of 1.5% agarose and 2.5μg/mL propidium iodide and imaged every 10 min for a total of 6 hours (n = 1).
Video 3
Time-lapse series of E. coli pSCrhaB2-CV::tas1tox pre-induction. Cells were spotted on an LB growth pad comprised of 1.5% agarose and 2.5μg/mL propidium iodide and imaged every 10 min for a total of 6 hours (n = 1).
Video 4
Time-lapse series of E. coli pSCrhaB2-CV::tas1tox 1-hr following Tas1tox expression. Cells were spotted on an LB growth pad comprised of 1.5% agarose and 2.5μg/mL propidium iodide and imaged every 10 min for a total of 6 hours (n = 1).
Rights and permissions
About this article
Cite this article
Ahmad, S., Wang, B., Walker, M.D. et al. An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678 (2019). https://doi.org/10.1038/s41586-019-1735-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1735-9
This article is cited by
-
Conservation and similarity of bacterial and eukaryotic innate immunity
Nature Reviews Microbiology (2024)
-
Bacterial type VI secretion system (T6SS): an evolved molecular weapon with diverse functionality
Biotechnology Letters (2023)
-
Biology and evolution of bacterial toxin–antitoxin systems
Nature Reviews Microbiology (2022)
-
A secreted effector with a dual role as a toxin and as a transcriptional factor
Nature Communications (2022)
-
Direct activation of a bacterial innate immune system by a viral capsid protein
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.