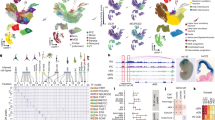
Abstract
The human brain has undergone substantial change since humans diverged from chimpanzees and the other great apes1,2. However, the genetic and developmental programs that underlie this divergence are not fully understood. Here we have analysed stem cell-derived cerebral organoids using single-cell transcriptomics and accessible chromatin profiling to investigate gene-regulatory changes that are specific to humans. We first analysed cell composition and reconstructed differentiation trajectories over the entire course of human cerebral organoid development from pluripotency, through neuroectoderm and neuroepithelial stages, followed by divergence into neuronal fates within the dorsal and ventral forebrain, midbrain and hindbrain regions. Brain-region composition varied in organoids from different iPSC lines, but regional gene-expression patterns remained largely reproducible across individuals. We analysed chimpanzee and macaque cerebral organoids and found that human neuronal development occurs at a slower pace relative to the other two primates. Using pseudotemporal alignment of differentiation paths, we found that human-specific gene expression resolved to distinct cell states along progenitor-to-neuron lineages in the cortex. Chromatin accessibility was dynamic during cortex development, and we identified divergence in accessibility between human and chimpanzee that correlated with human-specific gene expression and genetic change. Finally, we mapped human-specific expression in adult prefrontal cortex using single-nucleus RNA sequencing analysis and identified developmental differences that persist into adulthood, as well as cell-state-specific changes that occur exclusively in the adult brain. Our data provide a temporal cell atlas of great ape forebrain development, and illuminate dynamic gene-regulatory features that are unique to humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Sequence data that support the findings of this study have been deposited in ArrayExpress with the accession codes E-MTAB-7552 (single-cell RNA-seq data based on 10x Genomics), E-MTAB-8234 (single-cell RNA-seq data based on Fluidigm C1/Smart-seq2), E-MTAB-8089 (single-cell ATAC-seq of human organoids), E-MTAB-8043 (single-cell ATAC-seq of chimpanzee organoids), E-MTAB-8083 (single-cell ATAC-seq of bonobo organoids), E-MTAB-8087 (single-cell ATAC-seq of macaque organoids), E-MTAB-8228 (the bulk ATAC-seq data), E-MTAB-8230 (snRNA-seq data of the adult brain samples) and E-MTAB-8231 (bulk RNA-seq data). The expression data are also available for exploration in scApeX via the link https://bioinf.eva.mpg.de/shiny/sample-apps/scApeX/.
Code availability
The computational code used in this study is available at GitHub (https://github.com/quadbiolab/primate_cerebral_organoids) or upon request.
References
Sousa, A. M. M., Meyer, K. A., Santpere, G., Gulden, F. O. & Sestan, N. Evolution of the human nervous system function, structure, and development. Cell 170, 226–247 (2017).
Martin, R. D. Relative brain size and basal metabolic rate in terrestrial vertebrates. Nature 293, 57–60 (1981).
He, Z. et al. Comprehensive transcriptome analysis of neocortical layers in humans, chimpanzees and macaques. Nat. Neurosci. 20, 886–895 (2017).
Sousa, A. M. M. et al. Molecular and cellular reorganization of neural circuits in the human lineage. Science 358, 1027–1032 (2017).
Somel, M. et al. Transcriptional neoteny in the human brain. Proc. Natl Acad. Sci. USA 106, 5743–5748 (2009).
Konopka, G. et al. Human-specific transcriptional networks in the brain. Neuron 75, 601–617 (2012).
Zhu, Y. et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science 362, eaat8077 (2018).
Reilly, S. K. et al. Evolutionary genomics. Evolutionary changes in promoter and enhancer activity during human corticogenesis. Science 347, 1155–1159 (2015).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Marchetto, M. C. N. et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature 503, 525–529 (2013).
Camp, J. G. et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl Acad. Sci. USA 112, 15672–15677 (2015).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Velasco, S. et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 570, 523–527 (2019).
Amiri, A. et al. Transcriptome and epigenome landscape of human cortical development modeled in organoids. Science 362, eaat6720 (2018).
Mora-Bermúdez, F. et al. Differences and similarities between human and chimpanzee neural progenitors during cerebral cortex development. eLife 5, e18683 (2016).
Pollen, A. A. et al. Establishing cerebral organoids as models of human-specific brain evolution. Cell 176, 743–756 (2019).
Weinreb, C., Wolock, S. & Klein, A. M. SPRING: a kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics 34, 1246–1248 (2018).
Miller, J. A. et al. Transcriptional landscape of the prenatal human brain. Nature 508, 199–206 (2014).
Nowakowski, T. J. et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Renner, M. et al. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 36, 1316–1329 (2017).
Bakken, T. E. et al. A comprehensive transcriptional map of primate brain development. Nature 535, 367–375 (2016).
Leigh, S. R. Brain growth, life history, and cognition in primate and human evolution. Am. J. Primatol. 62, 139–164 (2004).
Marchetto, M. C. et al. Species-specific maturation profiles of human, chimpanzee and bonobo neural cells. eLife 8, e37527 (2019).
Otani, T., Marchetto, M. C., Gage, F. H., Simons, B. D. & Livesey, F. J. 2D and 3D stem cell models of primate cortical development identify species-specific differences in progenitor behavior contributing to brain size. Cell Stem Cell 18, 467–480 (2016).
Florio, M. et al. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. eLife 7, e32332 (2018).
Visel, A., Minovitsky, S., Dubchak, I. & Pennacchio, L. A. VISTA Enhancer Browser—a database of tissue-specific human enhancers. Nucleic Acids Res. 35, D88–D92 (2007).
Gregorio, I., Braghetta, P., Bonaldo, P. & Cescon, M. Collagen VI in healthy and diseased nervous system. Dis. Model. Mech. 11, dmm032946 (2018).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Kilpinen, H. et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature 546, 370–375 (2017).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Okita, K. et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31, 458–466 (2013).
Lancaster, M. A. & Knoblich, J. A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 9, 2329–2340 (2014).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Angerer, P. et al. destiny: diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016).
He, Z., Bammann, H., Han, D., Xie, G. & Khaitovich, P. Conserved expression of lincRNA during human and macaque prefrontal cortex development and maturation. RNA 20, 1103–1111 (2014).
Dougherty, M. L. et al. Transcriptional fates of human-specific segmental duplications in brain. Genome Res. 28, 1566–1576 (2018).
Fiddes, I. T. et al. Human-specific NOTCH2NL genes affect Notch signaling and cortical neurogenesis. Cell. 173, 1356–1369 (2018).
Dennis, M. Y. et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell 149, 912–922 (2012).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Buenrostro, J. D. et al. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523, 486–490 (2015).
Renaud, G., Stenzel, U. & Kelso, J. leeHom: adaptor trimming and merging for Illumina sequencing reads. Nucleic Acids Res. 42, e141 (2014).
Renaud, G., Stenzel, U., Maricic, T., Wiebe, V. & Kelso, J. deML: robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics 31, 770–772 (2015).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011).
Pliner, H. A. et al. Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871 (2018).
Preissl, S. et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci. 21, 432–439 (2018).
Suzuki, K. et al. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 15, 31–36 (2014).
Mayer, C. et al. Developmental diversification of cortical inhibitory interneurons. Nature 555, 457–462 (2018).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Lindblad-Toh, K. et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478, 476–482 (2011).
Prabhakar, S., Noonan, J. P., Pääbo, S. & Rubin, E. M. Accelerated evolution of conserved noncoding sequences in humans. Science 314, 786 (2006).
Gittelman, R. M. et al. Comprehensive identification and analysis of human accelerated regulatory DNA. Genome Res. 25, 1245–1255 (2015).
Cagan, A. et al. Natural selection in the great apes. Mol. Biol. Evol. 33, 3268–3283 (2016).
Peyrégne, S., Boyle, M. J., Dannemann, M. & Prüfer, K. Detecting ancient positive selection in humans using extended lineage sorting. Genome Res. 27, 1563–1572 (2017).
Prüfer, K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014).
Chintalapati, M., Dannemann, M. & Prüfer, K. Using the Neandertal genome to study the evolution of small insertions and deletions in modern humans. BMC Evol. Biol. 17, 179 (2017).
McLean, C. Y. et al. Human-specific loss of regulatory DNA and the evolution of human-specific traits. Nature 471, 216–219 (2011).
Fu, Y. et al. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 15, 480 (2014).
Lambert, S. A. et al. The human transcription factors. Cell 172, 650–665 (2018).
Lake, B. B. et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science 352, 1586–1590 (2016).
Lake, B. B. et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 36, 70–80 (2018).
Yu, Q. & He, Z. Comprehensive investigation of temporal and autism-associated cell type composition-dependent and independent gene expression changes in human brains. Sci. Rep. 7, 4121 (2017).
Hafemeister, C. & Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Preprint at bioRxiv https://doi.org/10.1101/576827 (2019).
Sloan, S.A. et al. Human astrocyte maturation captured in 3D cerebral cortical spheroids derived from pluripotent stem cells. Neuron 95, 779–790 (2017).
Acknowledgements
We thank D. Wollny, A. Brazovskaya, K. Köhler, T. Schaffer, B. Schellbach, A. Weihmann, R. Schultz, I. Bünger, M. Dannemann, R. Snabel, B. Vernot, W. Hevers, M. Schörnig, J. Kelso and K. Sekine for their help with this project; A. Fischer, M. Halbwax, K. Köhler and the Tchimpounga Sanctuary for support with generating the JoC iPSC line; L. Berninger and J. Peters for contributing Sc102a1 and SandraA organoids; B. B. Lake for sharing nuclei isolation protocols; and A. Fischer, M. Halbwax and the Lola ya Bonobo Sanctuary in Congo for contributing bonobo tissue for single-nucleus experiments. Karyotyping was supported by the Stem Cell Engineering Facility of CMCB at Technische Universität Dresden. Sorting was in part performed at the CUDZ at the Veterinary Medicine Faculty at the University of Leipzig. The silhouette images in the figures are credited to istockphoto.com for human (cole matt) and chimpanzee (A-digit), and flaticon.com for macaque (Freepik). This work was supported by the Max Planck Society (B.T.), Chan–Zuckerberg Initiative (B.T. and J.G.C.), European Research Council (Anthropoid, J.G.C.; Organomics; B.T.) and the NOMIS Foundation (S.P.). S.K. was supported by the Boehringer Ingelheim Fonds.
Author information
Authors and Affiliations
Contributions
S.K. and M.J.B. grew organoids with assistance from A.W., L.S. and M.H. S.K. performed scRNA-seq and snRNA-seq with assistance from M.S. M.J.B. performed scATAC-seq. Z.H., M.J.B. and S.K. analysed the data. F.S.C. and M.H. performed immunohistochemical stainings. J.S.F. compared organoid scRNA-seq data to mouse voxel maps. P.G. dissected and sliced tissue for snRNA-seq. D.H. and Z.Q. performed bulk RNA-seq of adult tissue. S.K., M.J.B., Z.H., B.T. and J.G.C. designed the study and wrote the manuscript with support from P.K., W.B.H. and S.P.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Christopher Walsh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
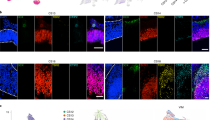
Extended Data Fig. 1 Differentiation and immunohistochemical characterization of human and chimpanzee cerebral organoids.
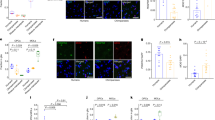
a, Phase contrast (PSC to neuroepithelium; scale bars, 200 μm; H9 for human, SandraA for chimpanzee) and bright-field images (organoid; scale bars, 1 mm; H9 and Wibj2 for human, JoC and SandraA for chimpanzee) showing examples of different stages of organoid development for human and chimpanzee. b, Immunohistochemical staining for PAX6 (green) and BCL11B (also known as CTIP2) (pink) of a 63-day human organoid from iPSC line 409b2 (left) and a 63-day chimpanzee organoid from iPSC line SandraA (right), with a magnified view into a cortical-like region (scale bars, 100 μm). c, Immunohistochemical staining of human (top left, Sc102a1, 50-day; top middle, 409b2, 63-day, the same organoid as the human organoid in b) and chimpanzee (bottom left, SandraA, 50-day; bottom middle, SandraA, 63-day) organoids (scale bars, 200 μm) for progenitor (PAX6) intermediate progenitor (TBR2) and deep-layer neurons (BCL11B) in whole organoids. Staining for deep-layer (BCL11B) and upper-layer (SATB2) neuron markers for human (top right, Sc102a1, 45-day) and chimpanzee (bottom right, SandraA, 63-day) organoids.
Extended Data Fig. 2 Heterogeneity analysis during human cerebral organoid development from pluripotency.
a, Cells from different human cell lines (23,226 cells from H9 and 20,272 cells from 409b2) were integrated using CCA and visualized using t-SNE. b, t-SNE coloured on the basis of cell line and batch. c, t-SNE coloured on the basis of time point. Heterogeneity analysis was performed on combined cells from day 0 of differentiation to 4-month-old organoids for iPSC and ESC-derived cells. d, Distribution of number of genes and UMIs for different time points and cell lines. e, Clustering was performed using the top-20 PCs as input for t-SNE and cluster names were assigned on the basis of expression of cluster marker genes and known marker genes. SC, stem cells; NEC, neuroectoderm-like cells; NSC, neural stem cells; g/oRGC, gliogenic/outer radial glia cells; G2M/S NPC, neural progenitor cells in G2M/S phase; G2M/S DP, dorsal progenitor cells in G2M/S phase; CN, cortical neurons; G2M/S vP, ventral progenitors in G2M/S phase; M/H, midbrain/hindbrain; CP, choroid plexus; M, mesenchymal-like cells. f, t-SNE plots coloured by expression level of selected marker genes on the basis of non-integrated expression values. g, Heat map showing averaged cluster expression for representative marker genes for clusters ordered according to their differentiation time from early to later stages and regional identity from dorsal to ventral forebrain and non-forebrain cells. h, In situ hybridization images from the Allen Developing Mouse Brain Atlas (available from https://developingmouse.brain-map.org/) showing expression of Foxg1, Neurod6 and Dlx5 in the mouse developing forebrain and human whole-trajectory SPRING plots coloured by the corresponding genes. i, Pseudotemporal expression of example genes marking different stages of development over the whole human cerebral organoid developmental trajectory. j, Umbrella plot showing the similarity of each organoid cell to a cell ‘prototype’ generated from a reference scRNA-seq cell atlas of the human fetal cortex19. k, Plots show the proportion of organoid cells per time point that match a reference prototype.
Extended Data Fig. 3 Analysis of human cerebral organoid single-cell transcriptomes from seven individuals.
a, scRNA-seq was performed on two-month-old cerebral organoids from one human ESC and six iPSC lines. b, All data (49,153 cells) were combined and cell heterogeneity was assessed using t-SNE with the top 20 PCs as the input. Cells are also coloured by marker gene expression and RSS. c, t-SNE plot with RSS against Brainspan fetal reference data as the input (RSS-t-SNE), coloured by cell lines. Cells from different lines are well integrated. d, SPRING plot of two-month-old human organoid pseudocells (9,650), coloured by neuronal trajectory branches and pseudotimes. e, SPRING plots of two-month-old human organoid cells, coloured by marker gene expression. f, SPRING plots coloured by cell line show contributions of each line to different branches of the trajectory. g, Correlations of expression trajectories of genes with pseudotime-dependent expression patterns between cortical cells from each line to the others (pink), ventral cells from each line to others (blue), and cortical and ventral cells from the same lines after or before aligning the cortical and ventral pseudotimes (purple). h, Spatial location inference of neuron subtypes in human cerebral organoids. Left, bar plots show proportion of cells of each cell type that show highest gene-expression-pattern similarity to the average expression patterns in different structures, on the basis of the processed in situ hybridization image data (E13.5) provided in the Developing Mouse Brain database of Allen Brain Atlas (available from https://developingmouse.brain-map.org/). Expression similarity was calculated based on highly variable genes of the scRNA-seq data (top) or regional markers defined with the in situ hybridization data (bottom). Right, correlation patterns of average regional marker gene expression of each neuron subtype to voxels in five example sections (E13.5), as well as the structural annotation of the sections. i, Expression of two marker genes of diencephalon inhibitory neurons (PCP4 and RSPO3) in the SPRING embeddings, and their spatial expression patterns in E13.5 mouse brain (data from Allen Brain Atlas, available from https://developingmouse.brain-map.org/).
Extended Data Fig. 4 Heterogeneity analysis during chimp cerebral organoid development from pluripotency.
a, Heterogeneity analysis for iPSC-derived chimpanzee cells (36,884) from day 0 of differentiation to 4 months of organoid development for one cell line (SandraA). b, Heat map visualizing averaged cluster expression for marker genes with columns ordered based on differentiation progress from early-to-late time points and regional identity sorted from dorsal to ventral forebrain to non-forebrain cells and non-ectodermal-derived cells. c, Cluster identification and t-SNE using the top-15 PCs for clustering. Cluster assignment was on the basis of cluster markers as well as expression patterns of known marker genes. SC, stem cells; G2M/S DP, dorsal progenitors in G2M/S phase; dlN, deep-layer neuron; ulN, upper-layer neurons; vP/N, ventral progenitor/neuron; M – mesenchymal-like cells; OL, off-lineage cells. d, t-SNE plots coloured on the basis of gene expression of representative marker genes used to assign cluster identities. e, SPRING plots of whole developmental trajectory for human and chimpanzee coloured by marker genes. f, Pseudotemporal gene-expression patterns showing marker genes for early, dorsal and ventral branches for human and chimpanzee. g, Schematic showing alignment of human and chimpanzee pseudotimes after combining pseudocells from the early stages and the dorsal forebrain lineage. The later chimpanzee pseudotime points fail to align with human pseudocells. h, Expression of genes duplicated in humans projected onto the human whole-lineage SPRING analysis, with the G2M/S phase marker MKI67 shown as a reference.
Extended Data Fig. 5 Analysis of cell-type heterogeneity of cerebral organoids and fetal cortical tissues based on scRNA-seq data from Fluidigm C1.
a, Overview of the Fluidigm C1 scRNA-seq data. Each dot represents a cerebral organoid or fetal brain sample from one cell line or species at a certain age, with its size showing the number of cells measured. The left panel shows organoid sample information as published in Pollen et al. (2019)16 (excluding redundant cells from Camp et al. (2015)11 and Mora-Bermudez et al. (2016)15), including the data initially published in Sloan et al. (2017)70. The middle panel shows organoid sample information generated in Camp et al. (2015)11, Mora-Bermudez et al. (2016)15 and in this study. The right panel shows fetal prefrontal cortex sample information reported in Nowakowski et al. (2017)19. b, All cerebral organoid data (5,838 cells) were combined and cell heterogeneity was assessed using t-SNE with the RSS profiles to the fetal Brainspan data as the input. Cells are coloured by cell type or cluster, species, institutions generating the data, dorsal trajectory pseudotimes and marker gene expression. c, t-SNE plots for all fetal brain data (5,080 cells) to assess cell heterogeneity, with the RSS profiles to the fetal Brainspan references as the input. Cells are coloured by cell type or cluster, species, dorsal excitatory neuron trajectory pseudotimes and marker gene expression. d, Heat map showing marker gene-expression patterns across different cell types in the droplet-based organoid scRNA-seq data generated in this manuscript and the above described C1-based scRNA-seq data.
Extended Data Fig. 6 Analysis of neuron maturation timing difference in human, chimpanzee and macaque cerebral organoids.
a, Box plots (IQR with minimum and maximum, outliers removed) showing sum expression levels (in reads per kilobase of transcript per million (RPKM)) of genes with GO annotation neuron projection (1,487 genes), neuron differentiation (1,367 genes), synapse assembly (168 genes) and neurotransmitter secretion (169 genes) in bulk RNA-seq data from Brainspan fetal cortical samples from 8 PCW to 17 PCW. b, Box plots showing sum expression levels of the same gene lists in fetal human dorsal excitatory neurons along the estimated developmental pseudotimes (Nowakowski et al. (2017) dataset19). c, Projection of human and chimpanzee organoid cells to human fetal brain data reveals higher similarity of chimpanzee organoid cells to later stages of development compared to human organoid cells. d, Box plots showing sum expression levels of genes with specific annotation to only one of the four GO terms in human and chimpanzee pseudocells (1,791 human and 4,304 chimp) along the cortical pseudotimes. Heat maps showing expression of example genes from the GO terms for human and chimp along pseudotime bins. The Venn diagram on the left shows the overlap of genes related to the four GO terms. e, Distribution of neuron projection scores of human and chimpanzee cortical cells (388 human and 355 chimp) reported in Pollen et al. (2019) along the cortical pseudotimes16. Each dot represents one cell, and is coloured by the organoid cell line. Light colours represent human cell lines and dark colours represent chimpanzee ones. Two-sided Wilcoxon’s rank-sum test (*P = 0.013 and **P = 0.004). f, Observed timing difference of upper-deeper layer specification in human and chimpanzee cerebral organoids from 10x Genomics data generated in this study. The left panel shows expression of cortical deep (BCL11B, left) and upper (SATB2, right) layer marker genes projected onto the chimpanzee (top) and human (bottom) SPRING plot. BCL11B and SATB2 become anti-correlated in their pseudotemporal expression profile in both human and chimpanzee (right), while the onset of anti-correlation happens earlier in chimpanzee than in human. g, Abundance of upper-layer neurons relative to deeper-layer neurons in human and macaque fetal prefrontal cortex samples19 in Nowakowski et al. (2017) grouped by early time points (<100 days old) or all time points combined. h, scRNA-seq was performed on two-to-four-month cerebral organoids from a macaque iPSC line. The SPRING plot of pseudocells (2,913) was constructed with the top-20 PCs as the input. The heterogeneity analysis suggests multiple cell types in the macaque organoids, including cortical neurons, NPCs, astrocytes and other cell types such as retina and mesenchyme-like cells. i, SPRING plot coloured by pseudotimes of cortical pseudocells, which are the pseudocells’ quantiles of diffusion component (DC) 1 of the cortical pseudocells diffusion map. j, SPRING plot coloured by marker gene expression. k, The onset of anti-correlation between SATB2 and BCL11B occurs earlier along the macaque pseudotime (1,107 pseudocells), relative to human (1,118 pseudocells) and chimpanzee (1,645 pseudocells), when focusing on the two-month cerebral organoids. l, Box plots (box shows IQR and whiskers show 1.5 × IQR) showing the neuron projection scores in human, chimpanzee and macaque along the unaligned cortical pseudotimes.
Extended Data Fig. 7 Pseudotime alignment between primates and differential expression between human and chimpanzee.
a, SPRING plots visualizing the kNN networks of human (10,063) and chimpanzee (5,612) pseudocells, and macaque cells (6,580), which represent NPCs and neurons of different brain regions. Cortical NPCs and neurons are coloured by their pseudotimes. b, Ratios of upper layer (UL) to deeper layer (DL) neuron marker expression in human (black), chimpanzee (dark grey) and macaque (light grey) organoids. The dashed line indicates the cut-off applied to human pseudocells to filter out those representing UL neurons. c, Truncated dynamic time warping (DTW)-based alignment was applied to align human, chimpanzee and macaque cortical pseudotime courses. Two support vector regression models were trained to predict chimpanzee (top) and macaque (bottom) pseudotimes of human pseudocells. A constrained B-splines regression model was fitted to determine the trimming point at the chimpanzee and macaque pseudotime courses, respectively. An end-to-end DTW-based alignment was applied to the human pseudotime course to the trimmed chimpanzee and macaque pseudotime courses for the final alignments (middle). d, Pseudotemporal expression profiles of GLI3, EOMES and BCL11B along the human, chimpanzee and macaque cortical pseudotimes, before (left) and after (right) the pseudotime alignment procedures. e, Robustness and false-positive rate of differential pseudotemporal expression between human and chimpanzee based on the number of cell lines involved in the analysis with constrained replaceable pseudocell subsampling. In each subsampling, pseudocells representing cells from a certain number of human lines were sampled in a replaceable manner to recapitulate pseudocell distribution along pseudotime course of the chimpanzee pseudocells. Differential expression analysis was applied to compare all chimpanzee pseudocells and the sampled human pseudocells to estimate robustness to cell line numbers (dark grey boxes), and to compare sampled human pseudocells to human pseudocells from another two lines sampled with the same procedure to estimate false-positive rate (light grey boxes). In box plots, boxes represent 100 times of subsampling IQR, the line represents 1.5 × IQR and dots represent outliers. f, Robustly detected human–chimpanzee differentially expressed genes (robust DE genes) are defined as the non-ribosomal genes which were detected as DE in at least 80% of the subsampling-based human–chimpanzee DE analysis using any number of human lines (black). The dendrogram shows the hierarchical clustering of robust DE genes, based on their human–chimpanzee pseudotemporal DE patterns along the aligned pseudotimes of cortical organoid pseudocells, resulting in eight clusters of robust DE genes. g, Pseudotemporal differential expression patterns between human and chimpanzee (without including macaque cells) of the eight clusters of genes along the pseudotimes of cortical organoid pseudocells with 50% and 95% confidence intervals shown in dark and light grey, respectively. Numbers of genes in each cluster are shown in parenthesis. h, Number of differentially expressed genes in chimpanzee versus human and macaque comparison grouped by gain or loss of expression in chimpanzees. A gain of expression specifically in chimpanzees is more likely than a loss of expression pattern conserved in the other primates. i, Comparison of the reported human–chimpanzee pseudotemporal differential expression based on 10x Genomics data with the Fluidigm C1-based scRNA-seq data of human and chimpanzee cerebral organoids. The two rows show the results based on C1 data generated in this manuscript and combined with data from refs. 11,15,16. The first two columns show estimated human–chimpanzee differential expression directionality and magnitude in the reported droplet-based scRNA-seq data and the C1-based measurement, with the first column presenting the generalized differential expression along the whole cortical pseudotimes, and the second column presenting the maximum differential expression along the pseudotimes. The red dots represent consistently differentially expressed genes, which have consistent differential expression directionalities in the two datasets. The right panel shows pseudotime intervals with the largest human–chimpanzee differential expression in the two datasets in comparison to the consistent differentially expressed genes. Dot sizes represent frequencies. j, Comparison of the estimated human–macaque differential expression directionality and magnitude of the human-specific differentially expressed genes using human and macaque fetal prefrontal cortex scRNA-seq data16,19. k, Functional annotations of genes with human-specific expression patterns based on GO annotations related to brain development and neurogenesis. Only the human-specific differentially expressed genes with consistent human–chimpanzee or human–macaque differential expression detected in at least one of the three C1-based scRNA-seq datasets are shown. l, Ventral telencephalon cell heterogeneity in organoids was investigated by t-SNE embeddings with RSS profiles of human (3,385) and chimpanzee ventral (773) pseudocells combined as the input. Pseudocell clusters were annotated on the basis of marker gene expression. Pseudocells were also coloured by species and diffusion map based on MGE neuron developmental pseudotimes. m, t-SNE plots coloured by marker gene expression and in situ hybridization images from the Allen Developing Mouse Brain Atlas (available from https://developingmouse.brain-map.org/) showing expression of Dlx5, Isl1 and Sox6 in the mouse developing ventral forebrain embryonic day 13.5 (E13.5). n, Human–chimpanzee ventral differentially expressed genes are largely shared along the dorsal forebrain developmental trajectories. o, Human–chimpanzee DE directionalities and magnitudes and DE gene detection rates on the two trajectories. DE directionalities and magnitudes are consistent on the dorsal and MGE trajectories, with most of the shared DE genes showing the highest human–chimpanzee expression divergence at NPC. DE genes specifically detected on one trajectory have the tendency of higher detection rates on the trajectory where human–chimpanzee differential expression is detected.
Extended Data Fig. 8 Chromatin accessibility in cerebral organoids during development.
a, t-SNE projection of highly variable gene expression in Fluidigm C1-based scRNA-seq data of cerebral organoids. Cortical cells are coloured red, with larger points corresponding to cells with paired expression and chromatin accessibility data (data generated from the same cell suspension). A total of 94.4% (219 out of 232 cells) of cells with paired data are cortical, validating the cortical origins of the dissected cerebral organoid regions. b, Cerebral organoid accessible peaks are significantly and highly enriched (Fisher’s exact test) for overlapping human VISTA enhancers active in the forebrain relative to all other tissues (left). Three representative human VISTA enhancers with validated activity in E11.5 mouse forebrain that overlap cerebral organoid peaks (out of 268 such enhancers) (right). c, Percentage of genes with accessible chromatin at the promoter of genes that are expressed or not expressed in human cerebral organoids. d–f, t-SNE projection of bias-corrected deviations in accessibility for 7-mers within organoid scATAC-seq peaks per cell, with cells coloured by cell state (NPC, neuron) for human (d, 221 cells), chimpanzee (e, 543 cells) and macaque (f, 118 cells). Binding motif deviation Z-scores for representative transcription factors are shown, as well as deviation Z-scores for overlapping DA snATAC-seq peaks in mouse developing forebrain excitatory neurons51. g, Signal intensity tracks of aggregated and individual single-cell chromatin accessibility data per cell state in human organoids at a NPC-specific promoter peak (left) and a neuron-specific promoter peak (right). For comparison, cerebral organoid bulk ATAC-seq chromatin accessibility data and human fetal brain bulk DNase-seq are shown. h, Enrichment of representative enriched biological process GO terms associated with human NPC DA peaks (gold) or human neuron DA peaks (light red) relative to all human organoid accessible peaks. Each point in the scatter plot represents a GO term and is coloured by their enrichment in NPCs (yellow), neurons (red), both (dark red) or neither (grey). i, t-SNE plots coloured by pseudotime, and heat maps showing binding motif deviation Z-scores for chosen transcription factors (rows) in all cells (columns) ordered in pseudotime for human (left) and chimpanzee (right). j, t-SNE projection of bias-corrected deviations in accessibility for 7-mers within scATAC-seq peaks per cell (518 cells), with cells coloured by time point, and organoid data coloured by cell state. Binding motif deviation Z-scores for representative transcription factors are shown to the right. k, t-SNE plot with cells coloured by their deviation Z-score for overlapping differentially accessible snATAC-seq peaks from mouse developing forebrain51 radial glia cells (left) or excitatory neurons (right). l, Diffusion map projection using the top-250 differentially accessible peaks per time point or cell state. The principle curve fit through the cells is shown as a black line. m, Proportion of cells scaled by row for each time point or cell state over pseudotime. n, Heat map representing the deviation Z-score of transcription factor motifs that significantly vary over the time course plotted for each cell across pseudotime.
Extended Data Fig. 9 Chromatin accessibility differences in human and chimpanzee cerebral organoids.
a, Signal intensity tracks of aggregated single-cell and bulk chromatin accessibility data from human, chimpanzee and macaque at a human-specific NPC-specific DA peak (left) and a human-specific neuron-specific DA peak (right). b, The eight most significant human–chimp organoid DA peaks containing a fixed SNC and accessible only in the cerebral organoid stage that overlap a VISTA human enhancer with validated activity in the developing mouse forebrain (out of 68 such cases). For each DA peak, the accessibility across pseudotime is shown for human and chimpanzee with heat maps depicting cells where the peak is accessible (yellow) or inaccessible (black). The activity pattern of the overlapping VISTA enhancer in E11.5 mouse embryos is shown to the right. c, The proportion of DE genes (dark colour) or all expressed genes as background (light colour) with a human–chimp organoid DA peak overlapping the promoter region (blue) or distal to the promoter region (pink). The plot shows that DE genes between human and chimpanzee organoids are more likely to have a nearby DA peak than background. d, Fixed SNCs predicted to significantly alter transcription factor binding within human–chimp organoid DA peaks, with the name of the altered motif shown for peaks linked to DE genes (red points). On the right, signal intensity tracks for a human motif gain (top) and human motif loss (bottom) within a human–chimp DA peak. e, Altered transcription factor motifs grouped by family plotted for their alteration rate, which is the number of times a family member’s motif is altered in human–chimp organoid DA peaks divided by the number of times it is detected in all accessible organoid peaks. f, Twenty transcription factors with the highest alteration rate, which is the number of times a motif is altered in human–chimp organoid DA peaks divided by the number of times it is detected in all accessible organoids peaks. Heat maps show their expression level in human and chimpanzee NPCs and neurons, with the bars to the left representing the average expression level across NPCs and neurons. g, Example of an accessible peak in chimpanzee and macaque that overlaps a computationally verified, non-polymorphic human conserved deletion (hCONDEL). h, i, Signal intensity tracks of aggregated single-cell or bulk chromatin accessibility data from human, chimpanzee and macaque for two genes, LYPD1 (h) and RAC1 (i), that have higher expression and specifically in humans, with genomic regions with gain of accessibility detected specifically in humans. Gene expression is shown in heat maps (bottom).
Extended Data Fig. 10 Supplementary analysis of human, chimpanzee and macaque adult brain snRNA-seq.
a, The snRNA-seq data of adult brains in human (50,035), chimpanzee and bonobo (33,847) and macaque (50,403) were integrated using Seurat v.3. b, Box plots (boxes show IQR and whiskers show 1.5 × IQR) showing the number of detected genes in single nuclei and pseudonuclei (3,420 human, 3,831 chimpanzee and 4,623 macaque pseudonuclei). c, Heat map showing the average prediction scores of each of the 20 identified clusters to each of the cell types reported66 by Lake et al. (2016), as well as their estimated distributions in different cortical layers in humans. Clusters are grouped in major cell classes. d, Cell-type composition of layers and layer distribution of cell types in human. Left, stacked bars showing the estimated cell-type composition of different layers. Right, box plots (boxes show IQR and whiskers show 1.5 × IQR) showing the estimated proportion per layer for four cell classes: excitatory neurons, inhibitory neurons, astrocytes and oligodendrocytes. e, Genomic conservation based on average phastCon scores of developmental stage markers (in total 2,000 genes) from iPSCs to neurons in human cerebral organoids (***P < 10−10, two-sided Wilcoxon’s rank-sum test, n1 = 818 genes, n2 = 188 genes). f, Hierarchical clustering of the average transcriptome of seven cell classes in the three species. g, Expression of layer markers (RASGRF2, RORB, ETV1 and TLE4) in excitatory neurons and inhibitory neuron subtype markers (PVALB and SST) in inhibitory neurons, along the predicted laminar origin of the pseudonuclei in human, chimpanzee and bonobo, and macaque. h, Detection rate in adult tissue of genes being differentially expressed between NPCs and neurons in organoids. i, Comparison of human–chimpanzee DE in adult excitatory neurons and that in organoid dorsal neurons for the robust DE genes detected in the organoid dorsal forebrain trajectory. Three categories of DE genes are highlighted: ubiquitous DE in organoids (top), DE only in NPCs (middle) and DE only in neurons (bottom). j, Comparison of gene-detection rates in organoid dorsal neurons and adult excitatory neurons, with human–chimpanzee DE genes in adult excitatory neurons coloured in yellow, DE genes in organoid dorsal neurons coloured in green, and shared DE genes coloured in black. The dashed curve shows the fitted relationship between the two systems using all genes. Area below the curve represents higher detection rate in organoid neurons than adult neurons and area above the curve represents higher detection rate in adult neurons. k, Comparison of human–chimpanzee DE (left) between organoid dorsal neurons and adult excitatory neurons, and between organoid ventral MGE neurons and adult inhibitory neurons (right). Densities are shown as grey scale shadows, with human–chimpanzee DE genes highlighted (yellow, DE only in adult; green, DE only in organoids; black, DE in both). l, Number of human and chimp DE genes for cell classes based on all cell types, a subset of cell types and specific cell types. m, Number of chimpanzee-specific DE genes across cell classes. The majority of the chimpanzee-specific DE genes have gain of expression (dark) rather than loss of expression (light).
Supplementary information
Supplementary Information
This file contains a guide to Supplementary Tables 1-15, Supplementary Notes and Supplementary References.
Supplementary Tables
This file contains Supplementary Tables 1-15 – see Supplementary Information document for guide.
Rights and permissions
About this article
Cite this article
Kanton, S., Boyle, M.J., He, Z. et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 574, 418–422 (2019). https://doi.org/10.1038/s41586-019-1654-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1654-9
This article is cited by
-
Single-nucleus RNA velocity reveals critical synaptic and cell-cycle dysregulations in neuropathologically confirmed Alzheimer’s disease
Scientific Reports (2024)
-
Human cytomegalovirus (CMV) dysregulates neurodevelopmental pathways in cerebral organoids
Communications Biology (2024)
-
A cell fate decision map reveals abundant direct neurogenesis bypassing intermediate progenitors in the human developing neocortex
Nature Cell Biology (2024)
-
Genetics of human brain development
Nature Reviews Genetics (2024)
-
A model of human neural networks reveals NPTX2 pathology in ALS and FTLD
Nature (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.