Abstract
During post-implantation development of the mouse embryo, descendants of the inner cell mass in the early epiblast transit from the naive to primed pluripotent state1. Concurrently, germ layers are formed and cell lineages are specified, leading to the establishment of the blueprint for embryogenesis. Fate-mapping and lineage-analysis studies have revealed that cells in different regions of the germ layers acquire location-specific cell fates during gastrulation2,3,4,5. The regionalization of cell fates preceding the formation of the basic body plan—the mechanisms of which are instrumental for understanding embryonic programming and stem-cell-based translational study—is conserved in vertebrate embryos6,7,8. However, a genome-wide molecular annotation of lineage segregation and tissue architecture of the post-implantation embryo has yet to be undertaken. Here we report a spatially resolved transcriptome of cell populations at defined positions in the germ layers during development from pre- to late-gastrulation stages. This spatiotemporal transcriptome provides high-resolution digitized in situ gene-expression profiles, reveals the molecular genealogy of tissue lineages and defines the continuum of pluripotency states in time and space. The transcriptome further identifies the networks of molecular determinants that drive lineage specification and tissue patterning, supports a role of Hippo–Yap signalling in germ-layer development and reveals the contribution of visceral endoderm to the endoderm in the early mouse embryo.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The RNA-seq data generated in this study were deposited in the NCBI Gene Expression Omnibus under accession number GSE120963 and the NODE project under accession number OEP000320. Our resources can be explored at the eGastrulation web portal (http://egastrulation.sibcb.ac.cn). All other data are available from the corresponding authors upon request.
Code availability
Custom codes for generating corn plots for different stages can be obtained from the eGastrulation web portal (http://egastrulation.sibcb.ac.cn).
Change history
16 September 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
03 January 2020
An Amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Smith, A. Formative pluripotency: the executive phase in a developmental continuum. Development 144, 365–373 (2017).
Lawson, K. A., Meneses, J. J. & Pedersen, R. A. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911 (1991).
Rivera-Pérez, J. A. & Hadjantonakis, A. K. The dynamics of morphogenesis in the early mouse embryo. Cold Spring Harb. Perspect. Biol. 7, a015867 (2014).
Tam, P. P. & Loebel, D. A. Gene function in mouse embryogenesis: get set for gastrulation. Nat. Rev. Genet. 8, 368–381 (2007).
Arnold, S. J. & Robertson, E. J. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 10, 91–103 (2009).
Irie, N. & Kuratani, S. The developmental hourglass model: a predictor of the basic body plan? Development 141, 4649–4655 (2014).
Tam, P. P. & Behringer, R. R. Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 68, 3–25 (1997).
Tam, P. P. & Quinlan, G. A. Mapping vertebrate embryos. Curr. Biol. 6, 104–106 (1996).
Griffiths, J. A., Scialdone, A. & Marioni, J. C. Using single-cell genomics to understand developmental processes and cell fate decisions. Mol. Syst. Biol. 14, e8046 (2018).
Kumar, P., Tan, Y. & Cahan, P. Understanding development and stem cells using single cell-based analyses of gene expression. Development 144, 17–32 (2017).
Pijuan-Sala, B. et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 566, 490–495 (2019).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Nowotschin, S. et al. The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367 (2019).
Peng, G. et al. Spatial transcriptome for the molecular annotation of lineage fates and cell identity in mid-gastrula mouse embryo. Dev. Cell 36, 681–697 (2016).
Boroviak, T. et al. Lineage-specific profiling delineates the emergence and progression of naive pluripotency in mammalian embryogenesis. Dev. Cell 35, 366–382 (2015).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Viotti, M., Nowotschin, S. & Hadjantonakis, A. K. SOX17 links gut endoderm morphogenesis and germ layer segregation. Nat. Cell Biol. 16, 1146–1156 (2014).
Fuxman Bass, J. I. et al. Using networks to measure similarity between genes: association index selection. Nat. Methods 10, 1169–1176 (2013).
Robb, L. & Tam, P. P. Gastrula organiser and embryonic patterning in the mouse. Semin. Cell Dev. Biol. 15, 543–554 (2004).
Balmer, S., Nowotschin, S. & Hadjantonakis, A. K. Notochord morphogenesis in mice: current understanding & open questions. Dev. Dyn. 245, 547–557 (2016).
Henrique, D., Abranches, E., Verrier, L. & Storey, K. G. Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875 (2015).
Kwon, G. S., Viotti, M. & Hadjantonakis, A. K. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Dev. Cell 15, 509–520 (2008).
Chan, M. M. et al. Molecular recording of mammalian embryogenesis. Nature 570, 77–82 (2019).
Nishioka, N. et al. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell 16, 398–410 (2009).
Wilson, V. & Beddington, R. S. Cell fate and morphogenetic movement in the late mouse primitive streak. Mech. Dev. 55, 79–89 (1996).
Briggs, J. A. et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018).
Peng, G., Tam, P. P. L. & Jing, N. Lineage specification of early embryos and embryonic stem cells at the dawn of enabling technologies. Natl. Sci. Rev. 4, 533–542 (2017).
Rivera-Pérez, J. A., Jones, V. & Tam, P. P. Culture of whole mouse embryos at early postimplantation to organogenesis stages: developmental staging and methods. Methods Enzymol. 476, 185–203 (2010).
Downs, K. M. & Davies, T. Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development 118, 1255–1266 (1993).
Chen, J. et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 12, 566–580 (2017).
Cui, G. et al. Spatio-temporal transcriptome construction of early mouse embryo with Geo-seq and Auto-seq. Protoc. Exch., https://doi.org/10.21203/rs.2.10081/v1 (2019).
Wells, J. M. & Melton, D. A. Early mouse endoderm is patterned by soluble factors from adjacent germ layers. Development 127, 1563–1572 (2000).
Liu, Q. et al. Lung regeneration by multipotent stem cells residing at the bronchioalveolar-duct junction. Nat. Genet. 51, 728–738 (2019).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Trapnell, C. et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 31, 46–53 (2013).
Langfelder, P. & Horvath, S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 (2008).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
de Hoon, M. J., Imoto, S., Nolan, J. & Miyano, S. Open source clustering software. Bioinformatics 20, 1453–1454 (2004).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017).
Klein, C. A. et al. Combined transcriptome and genome analysis of single micrometastatic cells. Nat. Biotechnol. 20, 387–392 (2002).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Chung, N. C. & Storey, J. D. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31, 545–554 (2015).
Zhang, W. et al. Integrating genomic, epigenomic, and transcriptomic features reveals modular signatures underlying poor prognosis in ovarian cancer. Cell Rep. 4, 542–553 (2013).
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T. & Salzberg, S. L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protocols 11, 1650–1667 (2016).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902 (2019).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Reimand, J. et al. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 44 (W1), W83–W89 (2016).
Weng, M. P. & Liao, B. Y. modPhEA: model organism Phenotype Enrichment Analysis of eukaryotic gene sets. Bioinformatics 33, 3505–3507 (2017).
Cabili, M. N. et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011).
Harrow, J. et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012).
Hong, F. et al. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22, 2825–2827 (2006).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Walker, M. G., Volkmuth, W., Sprinzak, E., Hodgson, D. & Klingler, T. Prediction of gene function by genome-scale expression analysis: prostate cancer-associated genes. Genome Res. 9, 1198–1203 (1999).
Peng, H., Ruan, Z., Long, F., Simpson, J. H. & Myers, E. W. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nat. Biotechnol. 28, 348–353 (2010).
Richardson, L., et al. EMAGE mouse embryo spatial gene expression database: (2014 update). Nucleic Acids Res. 42, D835–D844 (2014).
Acknowledgements
The authors thank Q. Zhou for critical discussions, Y. Chen and Y. Xu for technical support, and the chemical biology core facility and animal core facility in SIBCB. This work was supported in part by the ‘Strategic Priority Research Program’ of the Chinese Academy of Sciences (Grants, XDA16020501 to N.J., XDA16020404 to G.P., and XDA01010303 and XDB19020301 to J.-D.J.H.), National Key Basic Research and Development Program of China (2018YFA0107201 to G.P., 2018YFA0800100, 2018YFA0108000, 2017YFA0102700 and 2015CB964500 to N.J., and 2015CB964803 and 2016YFE0108700 to J.-D.J.H.), National Natural Science Foundation of China (31871456 to G.P., 31661143042, 91519314, 31630043, 31571513 and 31430058 to N.J., and 91329302, 31210103916 and 91749205 to J.-D.J.H.), Science and Technology Planning Project of Guangdong Province, China (2017B030314056), Frontier Research Program of Guangzhou Regenerative Medicine and Health Guangdong Laboratory (2018GZR110105013), Shanghai Natural Science Foundation (18ZR1446200) and Shanghai Municipal Commission for Science and Technology Grants (17411954900). P.P.L.T. is a NHMRC Senior Principal Research Fellow supported by the National Health and Medical Research Council of Australia (Research Fellowship grant 1110751).
Author information
Authors and Affiliations
Contributions
N.J. and G.P. conceived the study. N.J., G.P. and J.-D.J.H. supervised the project. N.J., P.P.L.T. and G.P. designed the experiments. G.P., G. Cui, J.C., S.C. and Z.L. performed the Geo-seq of embryos. Y.Q. performed animal husbandry. S.S. analysed the sequencing data and built the web portal. F.Y. conducted the embryonic stem cells and ex vivo perturbation experiments. R.W. helped to generate the pipeline for all germ-layer corn plot display. G. Chen helped with data analysis. G.P., P.P.L.T. and S.S. wrote the paper with the help of all other authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks Thorsten Boroviak, Anna-Katerina Hadjantonakis, Kyle Loh and Valerie Wilson for their contribution to the peer review of this work.
Extended data figures and tables
Extended Data Fig. 1 Geo-seq analysis.
a, Samples for Geo-seq: embryonic stages, biological replicates and the germ layers that were sampled for Geo-seq analysis. Stages: E5.5–E6.0, pre-gastrulation stage; E6.5, early streak; E7.0, late mid-streak; E7.5, no-bud stage. See Supplementary Table 1 for complete information. b, The strategy of sampling of cell populations in the epiblast–ectoderm and endoderm from E5.5 to E7.5 embryos and mesoderm in the E7.0 to E7.5 embryos (outlined in c). Samples were designated by the sequential (ascending) order of serial sections (1 for the most distal section) and the regions in the section (R, reference section; S, sample section; see histology images for orientation). For E5.5 embryo, the anterior–posterior axis has not yet been determined, so samples were denoted as Epi 1/2 and En 1/2. For E5.5–E7.5 embryos: Epi, epiblast; En, endoderm; A, anterior; P, posterior; AP, pooled from anterior and posterior; L, left side (1, anterior; 2, posterior); R, right side (1, anterior; 2, posterior); embryonic axes: anterior–posterior, A↔P; proximal–distal, P↔D; left–right L↔R. Scale bar, 50 µm. d, Box plot showing the number of detected genes (FPKM > 0) for samples of E5.5 to E7.5 embryos (E5.5: n = 10; E6.0: n = 18; E6.5: n = 30; E7.0: n = 73; E7.5: n = 83). The centre line marks the median and box edges represent 25th and 75th percentiles. e, Gene-expression density plot of Geo-seq data of samples from E5.5 to E7.5 embryo; the solid line is the mean value and shaded region represents the s.d. f, Corn plots showing the expression of representative germ-layer markers (ectoderm: Sox2; endoderm: Sox17; mesoderm: Mesp1; visceral endoderm: Hnf4a) at E5.5 to E7.5 stages. Similar results were obtained from embryo replicates. g, The expression of anterior visceral endoderm (AVE) and distal visceral endoderm (DVE) markers (Cer1, Dkk1 and Lefty1) at E5.5 and E6.0 stages. Similar results were obtained from embryo replicates. h, The comparison of digitally rendered gene-expression pattern and WISH results of seven examples of genes in the epiblast and endoderm at E6.5 stage. WISH images were obtained from the EMAP eMouse Atlas Project (www.emouseatlas.org)56. Embryos are oriented with posterior to the right and distal to the bottom. i, The comparison of the expression of selected coding and noncoding RNAs by Geo-seq and WISH in E6.5 epiblast (Crb3, Car2), E7.0 (Gm37335, RP23-458G12.2) and E7.5 ectoderm (Hoxb3os, A930024E05Rik). n > 3 for each gene. Spearman correlation plots of WISH and Geo-seq results are shown on the right. Embryos are oriented with posterior to the right and distal to the bottom.
Extended Data Fig. 2 Functional analysis of lncRNA.
a, Experimental design for tracking the expression of lncRNAs during in vitro differentiation of mouse embryonic stem cells (mESCs). b, c, Expression of anterior-specific (b) and posterior-specific lncRNAs (c), assayed by real-time PCR, during in vitro differentiation (days 0–6) of mouse embryonic stem cells (n = 3 for KSR and FBS). d, Experimental design for assessing the effect of the loss of lncRNA function on the differentiation of mouse embryonic stem cells in vitro. e, Ablation of lncRNA 3110004A29Rik led to downregulation of mesoderm genes: T, Eomes and Mesp1 at days 5 and 6 of culture (n = 3). f, Ablation of lncRNA RP23-458G12.2 disrupted the expression of mesendoderm genes: Pcdh7, Eomes, Mesp1 and Hand1 at later time points of the seven-day period of differentiation in vitro (n = 3). Gene expression was assayed by real-time PCR. Data are mean ± s.e.m. Significant difference by two-sided t-test; *P < 0.05, **P < 0.01 and ***P < 0.001.
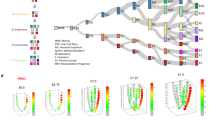
Extended Data Fig. 3 The segregation of germ-layer tissues by the regulon and transcriptome.
a, The workflow of regulon analysis in the SCENIC pipeline. Tissue samples from embryos at E2.5 to E7.5 were collated for mining the co-expression genes, cis-regulatory motif analysis and building of regulon units comprising of transcription factors (TF) and targets (TG), and the regulon-activity matrix for each sample were used in subsequent bioinformatic analysis (Methods). Dim, dimension. Transcriptome data of blastomeres of the morula, inner cell mass of the blastocyst and the primitive endoderm and epiblast of E4.5 embryo were sourced from ArrayExpress accession numbers E-MTAB-2958 and E-MTAB-2959. b, Corn plots show the spatial domain of the cell populations of each branch of the phylogenetic tree at E5.5–E7.5. c, t-SNE plots of tissue samples from E5.5 to E7.5 based on gene expression and the regulon-activity matrix (n = 214). d, e, PCA and t-SNE analysis for regulon-activity scores in the epiblast–ectoderm and mesoderm (n = 145) (d) and the primitive streak, mesoderm and endoderm of E5.5 to E7.5 embryos (n = 122) (e). f, Hierarchical clustering analysis for the posterior epiblast/primitive streak, the mesoderm and the endoderm for embryonic samples from E5.5 to E7.5 stages. g, The regulon-activity heat map for primitive streak, mesoderm and endoderm of E5.5 to E7.5 embryo. Sample in each column was ordered based on f, and examples of regulon transcription factors were listed. Regulon transcription factors coloured red were experimentally tested in this study. h, Corn plots showing the pattern of averaged activities of regulon groups in the endoderm, mesoderm and primitive streak samples.
Extended Data Fig. 4 The regulon sample gene sets.
a, t-SNE distribution of tissue samples of E2.5 to E7.5 embryos based on regulon-activity scores (n = 226). b, Two-dimensional PCA plots (n = 226) and corn plots showing the averaged regulon activities of G2, G3, G4, G6, G7, G8 and G9 regulon groups. c, The CSI co-expression network analysis (edges were filtered by CSI >0.85) for the nine regulon groups. Edges: positive interaction (brown), negative interaction (green). Triangle nodes denote transcription factors with knockout gastrulation phenotype in MGI database. A wider edge signifies higher correlation. d, The gene-expression profile of lineage markers in the different regulon sample groups.
Extended Data Fig. 5 Functional analysis of Sp1 of the G7 (pre-gastrulation), Hmgb3 of the G1 (late mesoderm/primitive streak) regulon group and Hmga2 of G8 (mesoderm) regulon group on differentiation of the mouse embryonic stem cells.
a, DNA sequence of two Sp1-knockout cell lines. b, Loss of Sp1 function led to downregulation of the naive marker genes (Rex1, Esrrb and Nanog) and upregulation of formative marker genes (Sall2, Fgf5 and Pou3f1) (n = 3). c, Loss of Hmgb3 function led to upregulation of Cdx2, Cdx4, Hoxa1 and Tbx6 genes related to precursors of the trunk mesoderm and no changes to markers of mesendoerm progenitors. Gene expression was assayed by qPCR. Data are mean ± s.e.m. (n = 3). Significant difference by two-sided t-test; *P < 0.05, **P < 0.01 and ***P < 0.001. d, DNA sequence and sequencing peak map of two Hmga2-knockout cell lines. e, Knockout of Hmga2 led to down regulation of cardiac progenitor marker genes at differentiation day 6 (n = 3). f, Knockout of Hmga2 led to down regulation of cardiomyocyte marker genes at differentiation day 12 (n = 3). g, Immunostaining of embryoid body at day 12 shows lower expression of cardiomyocyte marker c-TnT in Hmga2-knockout cells (scale bars: top, 25 μm; bottom, 5 μm) (n = 3). Data are mean ± s.e.m. Significant difference by two-sided t-test; *P < 0.05, **P < 0.01 and ***P < 0.001.
Extended Data Fig. 6 The spatial domains in the germ layers of E5.5–E7.0 embryos delineated by the DEG groups.
a–c, Heat maps and corn plots of DEG groups in E5.5 (a), E6.0 (b) and E6.5 (c) embryos. d, The transcriptional endoderm domains at E6.0 and E6.5 identified by analysis based on guilt-by-association co-expressed genes of Dkk1, Cer1 and Lefty1. e, Heat map and corn plots of DEG groups in E7.0 embryos. Representative genes of each gene group are listed. Corn plots show the spatial domains and the average expression level of the DEG groups. f, The activity score of Sox2 and T in the E7.0 primitive streak region.
Extended Data Fig. 7 The tissue spatial domains in the germ layers of E7.5 embryo and endoderm single-cell proportion in the endoderm.
a, Heat map and corn plots of DEG groups at the E7.5 stage. Representative genes of each DEG group are listed. Corn plots show the spatial domains and the average expression level of the gene group. b, Genes that are specifically expressed in the node region (DEG G5) of E7.5 embryos with high expression values and the corresponding JSD specificity score (in the distal-endoderm domain). c, Top ten (with the highest JSD scores) examples of node-specific expression pattern shown in corn plots. d, The dynamic expression of three E7.5 G5 genes during gastrulation shown in corn plots and compared with WISH results at E7.5 stage. Embryos are oriented with posterior to the right and distal to the bottom (n > 3 for each gene). e, Gene-filter criteria of single-cell RNA-seq data (n = 384 cells; top and bottom of each violin plot represent maxima and minima). Cells with number of detected genes less than 8,000 were discarded (left, red dotted line) and cells with the percentage of reads mapped to mitochondrial genes less than 0.2 were reserved for further analysis (right, red dotted line). f, UMAP plots of 345 single cells (that passed the quality check) in three clusters, indicted by different colours. g, Violin plots of gene-expression level of Sox17 and T across three clusters (SC-C1, n = 213 cells; SC-C2, n = 96 cells; SC-C3, n = 36 cells; top and bottom of each violin plot represent maximum and minimum, respectively). h, Feature plots of represented lineage markers (n = 345 cells). The colour gradient indicates levels of expression.
Extended Data Fig. 9 Signalling activity in the post-implantation embryo.
a, Enrichment for target and response genes of signalling pathways in the nine regulon gene groups (from Fig. 2a). Brown shows activating (A) and blue shows inhibitory (I) signalling activity. The significance of −log10(FDR) value in each cell was calculated by one-sided Fisher’s exact test followed by Benjamini–Hochberg correction. b, Corn plots showing the activity scores of the target and response genes related to the activated and inhibitory activity of BMP, Wnt, FGF, Nodal and Notch pathways in all germ-layer samples of E5.5 to E7.5 embryos. Similar results were obtained from embryo replicates. c, The expression of four Hippo–Yap signalling pathway components shown in corn plots of E7.5 embryo and in the dissected germ-layer tissues assayed by qPCR (n = 3). Dissected tissues: A, anterior epiblast, P, posterior epiblast, M, mesoderm; E, endoderm. d, WISH of E7.0 embryo (left) and visualization of hybridization signals in the germ layers (right) of Hippo pathway components Tead1 and Tead4, and Ctgf, the Yap1 downstream target gene (n > 3 for each gene). Dotted lines indicate plane of sectioning. e, The experimental design of ex vivo culture and treatment by inhibitor. E7.0 endoderm layer was explanted, treated with or without Yap inhibitor (verteporfin, VP) in N2B27 for 8 h and prepared for cDNA amplification and Geo-seq analysis. ExE, extra-embryonic tissues. f, g, The expression of Hippo–Yap pathway factors (Ctgf, P = 4.56 × 10−2; Cyr61, P = 3.38 × 10−2 and Vim, P = 8.8 × 10−3) (f) and visceral endoderm markers (Afp, P = 1.4 × 10−2; Amn, P = 1.2 × 10−3 and Ttr, P = 3.6 × 10−3) (g) following verteporfin treatment of endoderm explants. Data are mean ± s.e.m. Control, n = 10; verteporfin, n = 9. Significant difference by two-sided t-test; *P < 0.05 and **P < 0.01. h, Changes in the G3 regulon-activity scores following Yap inhibitor treatment in endoderm explants (regulon-activity scores were z-score-normalized across all verteporfin and control samples). Data are mean ± s.d. One-tailed t-test; P = 5.1 × 10−3; control, n = 3; verteporfin, n = 2. i, No effect on definitive endoderm genes (Sox17, Foxa2, Foxg1 and Pax9) after 8 h Yap inhibition of endoderm tissues ex vivo. Data are mean ± s.e.m. wild type, n = 10; verteporfin, n = 9.
Extended Data Fig. 10 The molecular fates and correlation with experimental cell fate maps.
a, Heat map showing the expression pattern of germ-layer marker genes. Top coloured bars indicate the developmental stage (stages: cyan, E6.5; yellow, E7.0; dark red, E7.5) and the location of the cell population in epiblast–ectoderm (region: red, anterior; blue, posterior; green, left; light green, right). The lineage fates (left bar) were assigned on the basis of the expression of markers of neuroectoderm (orange), surface ectoderm (yellow), mesoderm (green) and endoderm (red). b, Corn plots showing the activity scores of different lineage markers in embryos at different stages. c, (i) Corn plots showing the relative level of expression of germ-layer-related genes (as pie charts in the corn plots) in different cell populations of the epiblast–ectoderm in E6.5–E7.5 embryos; (ii) the molecular fate maps of major germ-layer derivatives in E6.5–E7.5 epiblast–ectoderm constructed based on the expression pattern of tissue markers; and (iii) the prospective fate maps (shown for the right lateral half of the epiblast–ectoderm) constructed from the data of the fate-mapping and lineage tracking experiments. Abbreviations: ne, neural ectoderm; fb, forebrain; mb, midbrain; hb, hindbrain; sc, spinal cord; se, surface ectoderm; am, amnion ectoderm; em, embryonic mesoderm; exm, extra-embryonic mesoderm; ht, heart; lm, lateral mesoderm; meso, mesoderm; pxm, paraxial mesoderm; end, endoderm; N, node; nd, notochord. d, Average expression of genes associated with epithelial–mesenchymal transition (EMT) showing enrichment of genes associated with enhanced EMT in the posterior epiblast (EMT up) and reduced EMT in the anterior epiblast (EMT down) in E6.5 to E7.5 embryos.
Supplementary information
41586_2019_1469_MOESM2_ESM.xlsx
Supplementary Table Supplementary Table 1: Complete sample information for all the developmental Geo-seq analysis. Stage, captured position and sample size were included.
41586_2019_1469_MOESM3_ESM.txt
Supplementary Table Supplementary Table 2: All the regulon values for the samples including Geo-seq data and published E2.5, E3.5, E4.5 data.
41586_2019_1469_MOESM4_ESM.xlsx
Supplementary Table Supplementary Table 3: The regulon unit (by TFs and targets) composition identified in the E2.5-E7.5 samples.
41586_2019_1469_MOESM5_ESM.xlsx
Supplementary Table Supplementary Table 4: The TF regulon networks that showed knockout phenotype in mouse genome information (MGI) database.
41586_2019_1469_MOESM6_ESM.xlsx
Supplementary Table Supplementary Table 5: The differentially expressed genes identified in respective stages from E5.5 to E7.5.
41586_2019_1469_MOESM7_ESM.xlsx
Supplementary Table Supplementary Table 6: The average expression (by means of FPKM) of spatial domains identified by DEGs from E5.5 to E7.5.
41586_2019_1469_MOESM10_ESM.xlsx
Supplementary Table Supplementary Table 9: All the primers (for qPCR test and in vitro probe generation) and gRNAs used in this study.
41586_2019_1469_MOESM11_ESM.wmv
Video 1: Hmga2 is critical for cardiac differentiation. Spontaneously contracting embryoid bodies (EBs) determined on day 12 of EB differentiation in Hmag2 control EBs.
41586_2019_1469_MOESM12_ESM.wmv
Video 2: Hmga2 is critical for cardiac differentiation. Spontaneously contracting EBs determined on day 12 of EB differentiation in Hmag2 knockout EBs.
Rights and permissions
About this article
Cite this article
Peng, G., Suo, S., Cui, G. et al. Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 572, 528–532 (2019). https://doi.org/10.1038/s41586-019-1469-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1469-8
This article is cited by
-
eSPRESSO: topological clustering of single-cell transcriptomics data to reveal informative genes for spatio–temporal architectures of cells
BMC Bioinformatics (2023)
-
Base editing-mediated one-step inactivation of the Dnmt gene family reveals critical roles of DNA methylation during mouse gastrulation
Nature Communications (2023)
-
Single cell RNA analysis of the left–right organizer transcriptome reveals potential novel heterotaxy genes
Scientific Reports (2023)
-
Three-dimensional molecular architecture of mouse organogenesis
Nature Communications (2023)
-
Time space and single-cell resolved tissue lineage trajectories and laterality of body plan at gastrulation
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.