Abstract
We sought to determine whether pre-eclampsia, spontaneous preterm birth or the delivery of infants who are small for gestational age were associated with the presence of bacterial DNA in the human placenta. Here we show that there was no evidence for the presence of bacteria in the large majority of placental samples, from both complicated and uncomplicated pregnancies. Almost all signals were related either to the acquisition of bacteria during labour and delivery, or to contamination of laboratory reagents with bacterial DNA. The exception was Streptococcus agalactiae (group B Streptococcus), for which non-contaminant signals were detected in approximately 5% of samples collected before the onset of labour. We conclude that bacterial infection of the placenta is not a common cause of adverse pregnancy outcome and that the human placenta does not have a microbiome, but it does represent a potential site of perinatal acquisition of S. agalactiae, a major cause of neonatal sepsis.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The 16S rRNA gene sequencing datasets generated and analysed in this study are publicly available under European Nucleotide Archive (ENA) accession number ERP109246. The metagenomics datasets, which primarily contain human sequences, are available with managed access in the European Genome-phenome Archive (EGA) accession number EGAD00001004198.
Change history
01 October 2019
An Amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Brosens, I., Pijnenborg, R., Vercruysse, L. & Romero, R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am. J. Obstet. Gynecol. 204, 193–201 (2011).
Aagaard, K. et al. The placenta harbors a unique microbiome. Sci. Transl. Med. 6, 237ra65 (2014).
Antony, K. M. et al. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am. J. Obstet. Gynecol. 212, 653.e1–653.e16 (2015).
Collado, M. C., Rautava, S., Aakko, J., Isolauri, E. & Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 6, 23129 (2016).
Perez-Muñoz, M. E., Arrieta, M. C., Ramer-Tait, A. E. & Walter, J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5, 48 (2017).
Salter, S. J. et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 12, 87 (2014).
Jervis-Bardy, J. et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome 3, 19 (2015).
de Goffau, M. C. et al. Recognizing the reagent microbiome. Nat. Microbiol. 3, 851–853 (2018).
Lauder, A. P. et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4, 29 (2016).
Leiby, J. S. et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 6, 196 (2018).
Theis, K. R. et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am. J. Obstet. Gynecol. 220, 267.e1–267.e39 (2019).
Leon, L. J. et al. Enrichment of clinically relevant organisms in spontaneous preterm delivered placenta and reagent contamination across all clinical groups in a large UK pregnancy cohort. Appl. Environ. Microbiol. 84, e00483-e18 (2018).
Sovio, U., White, I. R., Dacey, A., Pasupathy, D. & Smith, G. C. S. Screening for fetal growth restriction with universal third trimester ultrasonography in nulliparous women in the Pregnancy Outcome Prediction (POP) study: a prospective cohort study. Lancet 386, 2089–2097 (2015).
Hornef, M. & Penders, J. Does a prenatal bacterial microbiota exist? Mucosal Immunol. 10, 598–601 (2017).
Leong, H. N. et al. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J. Med. Virol. 79, 45–51 (2007).
Ravel, J. et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108 (suppl. 1), 4680–4687 (2011).
Glaser, P. et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 46, 1499–1513 (2002).
Abele-Horn, M., Scholz, M., Wolff, C. & Kolben, M. High-density vaginal Ureaplasma urealyticum colonization as a risk factor for chorioamnionitis and preterm delivery. Acta Obstet. Gynecol. Scand. 79, 973–978 (2000).
Schrag, S. J. et al. Group B streptococcal disease in the era of intrapartum antibiotic prophylaxis. N. Engl. J. Med. 342, 15–20 (2000).
Pasupathy, D. et al. Study protocol. A prospective cohort study of unselected primiparous women: the pregnancy outcome prediction study. BMC Pregnancy Childbirth 8, 51 (2008).
Gardosi, J., Mongelli, M., Wilcox, M. & Chang, A. An adjustable fetal weight standard. Ultrasound Obstet. Gynecol. 6, 168–174 (1995).
American College of Obstetricians and Gynecologists & Task Force on Hypertension in Pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet. Gynecol. 122, 1122–1131 (2013).
Lager, S. et al. Detecting eukaryotic microbiota with single-cell sensitivity in human tissue. Microbiome 6, 151 (2018).
Walker, A. W. et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3, 26 (2015).
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Nurk, S. et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 20, 714–737 (2013).
Johnson, M. et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9 (2008).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Carver, T., Harris, S. R., Berriman, M., Parkhill, J. & McQuillan, J. A. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469 (2012).
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K. & Schloss, P. D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120 (2013).
Eren, A. M. et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol. Evol. 4, 1111–1119 (2013).
Schmieder, R. & Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Ludwig, W. et al. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 (2004).
Viera, A. J. & Garrett, J. M. Understanding interobserver agreement: the kappa statistic. Fam. Med. 37, 360–363 (2005).
Mackinnon, A. A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement. Comput. Biol. Med. 30, 127–134 (2000).
Acknowledgements
The work was supported by the Medical Research Council (UK; MR/K021133/1) and the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre (Women’s Health theme). We thank L. Bibby, S. Ranawaka, K. Holmes, J. Gill, R. Millar and L. Sánchez Busó for technical assistance during the study. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Author information
Authors and Affiliations
Contributions
G.C.S.S., D.S.C.-J., J.P. and S.J.P. conceived the experiments. G.C.S.S., D.S.C.-J., J.P., S.J.P. and S.L. designed the experiments. S.L. and M.C.d.G. optimized the experimental approach. S.L. and F.G. performed the experiments. M.C.d.G. analysed all of the sequencing data. U.S. matched cases and controls, performed statistical analyses and provided logistical support for patient and sample metadata. E.C. managed sample collection and processing and the biobank in which all sample were stored. All authors contributed in writing the manuscript and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
J.P. reports grants from Pfizer, personal fees from Next Gen Diagnostics, outside the submitted work; S.J.P. reports personal fees from Specific, personal fees from Next Gen Diagnostics, outside the submitted work; D.S.C.-J. reports grants from GlaxoSmithKline Research and Development, outside the submitted work and non-financial support from Roche Diagnostics, outside the submitted work; G.C.S.S. reports grants and personal fees from GlaxoSmithKline Research and Development, personal fees and non-financial support from Roche Diagnostics, outside the submitted work; D.S.C.-J. and G.C.S.S. report grants from Sera Prognostics, non-financial support from Illumina, outside the submitted work. M.C.d.G., S.L., U.S., F.G. and E.C. have nothing to disclose.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peer review information Nature thanks David N. Fredricks and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Extended data figures and tables
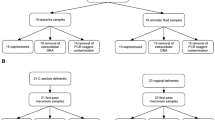
Extended Data Fig. 1 Two cohorts of placental samples were analysed.
Cohort 1 (n = 80) contained only samples from pre-labour Caesarean section (CS) deliveries and S. bongori was added to the samples before DNA isolation as a positive control. Samples in cohort 1 were analysed by both metagenomics and16S rRNA amplicon sequencing. Cohort 2 (n = 498) contained placental samples from Caesarean section and vaginal deliveries. DNA was isolated twice from each placental sample with two different DNA extraction kits. Samples were analysed by 16S rRNA amplicon sequencing. Pre-eclampsia (PE) was defined using The American College of Obstetricians and Gynaecologists (ACOG) 2013 definition. Small for gestational age (SGA) was defined as a birth weight less than the fifth percentile using a customized reference. Preterm denotes birth before 37 weeks gestation.
Extended Data Fig. 2 Positive control experiment comparison between metagenomics and 16S amplicon sequencing.
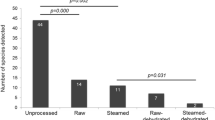
a, b, Adding approximately 1,100 CFUs of S. bongori to the placental tissue before DNA isolation resulted in an average of 180 reads (s.d. 90 reads) by metagenomic sequencing (n = 80) (a) or on average of 54% of all 16S rRNA amplicon sequencing reads (approximately 33,000 reads) being identified as S. bongori (s.d. 13%; n = 79) (b). Box represents the interquartile range; whiskers represent the maximum and minimum values; centre lines denote the median.
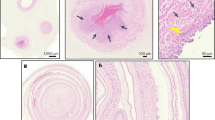
Extended Data Fig. 3 Strain analysis of E. coli reads found by metagenomics.
All reads identified in all 80 samples by Kraken25 as E. coli were extracted and mapped together against the closest E. coli reference genome (GenBank: CP02409.1). Single nucleotide polymorphisms, shown in red, were consistent for all samples across the genome. Single nucleotide polymorphisms were rare, except in the fimbrial chaperone protein gene (EcpD) indicated in light red. Sequence differences that appear as short sporadic red lines represent sequencing errors. Strain variation would have resulted in dashed vertical lines.
Extended Data Fig. 4 Detailed heat map metagenomic data.
Heat map showing the abundance of all non-human reads as detected by metagenomics. Human reads remaining after filtering (89.8%; s.d. 1.5%) are not shown for scaling purposes. Most taxa (shown on the right) are found in higher abundance within groups 1 and/or 2 (indicated on the left with light blue and purple, respectively). The purple box highlights the samples and species associated with group 2. The lane ID of each sample is represented by the first number (x axis). All samples from lanes 4 and 5 form group 2, and all samples from lanes 8 and 9 form group 3 (see Fig. 1a, b).
Extended Data Fig. 5 Species associated with batch effects visualized by PCA also do not show signal reproducibility.
a, PCAs of selections of samples from cohort 2 (16S), or of all cohort 2 samples as shown here, allows for the identification of batch effects and allows for the identification of contaminating species associated with the use of specific DNA isolation methods, kits and/or other reagents. An analysis of all samples shows that principal components 3 (x axis) and 4 (y axis) are strongly correlated with the use of Qiagen or specific Mpbio DNA isolation kits. b, Examples of bacteria detected in high abundance and frequency when processed with the Qiagen (x axis) and/or Mpbio (y axis) DNA isolation kits. Patterns that lack positive correlation (compare with Fig. 2a) demonstrate that signals are not sample- but batch-associated.
Extended Data Fig. 6 Scatterplot representations of the abundance of Bradyrhizobium, Burkholderia, vaginal lactobacilli and vaginosis bacteria during 16S amplicon sequencing.
a, b, The abundance of Bradyrhizobium (a) or Burkholderia (b) with respect to sequencing run batch effects during 16S amplicon sequencing. Numbers in parentheses indicate the number of samples sequenced in a given run. Values of zero are not shown on the logarithmic axis. c, d, The abundance of vaginal lactobacilli (c) and vaginosis bacteria (d) with respect to the mode of delivery during 16S amplicon sequencing. *P < 0.05, ***P < 0.001, Mann–Whitney U-tests, where values below 1% are regarded as 0% (not biologically relevant).
Extended Data Fig. 7 Mode of delivery and the detection of bacterial signals.
a, b, The association of vaginal lactobacilli with the mode of delivery, as determined by the analysis of 466 samples by 16S amplicon sequencing that were successfully sequenced twice using the Mpbio (a) and Qiagen (b) DNA isolation methods. Comparisons of the Mpbio and Qiagen DNA isolation techniques highlight that the same patterns are observed in the associations with mode of delivery. Comparisons also show that the Qiagen DNA isolation was more sensitive, resulting in twice as many signals above the 1% threshold. c–h, The association of bacterial groups with mode of delivery. Analyses were performed using all 498 placental samples with the highest value of either DNA isolation method for each bacterial group per sample. c, d, S. agalactiae was not associated with the mode of delivery irrespective of whether a 1% threshold was used (the minimum percentage considered to be potentially ecologically relevant) (c) or a 0.1% threshold was used (the 16S detection limit, relevant for detecting traces of contamination during delivery) (d). e, f, The Ureaplasma genus was significantly associated with the mode of delivery using the 0.1% threshold, similar to Fig. 2c, which describes the combination of all vaginosis-associated bacteria. g, h, F. nucleatum was not associated with the mode of delivery, irrespective of whether a 1% (g) or 0.1% (h) threshold was used. *P < 0.05, **P < 0.01, ***P < 0.001, Mann–Whitney U-tests.
Extended Data Fig. 8 Heat map of Spearman’s rho correlation coefficients of bacterial signals as found by 16S rRNA amplicon sequencing.
Sample-associated signals (red bar), are typically identified by increased kappa scores, as shown in Supplementary Table 4. Reagent contaminants are indicated by a blue bar. Vaginosis-associated bacteria (purple bar) show positive correlations (purple square) with each other, Lactobacillus iners and faecal bacteria (brown bar). Lactobacilli (yellow bar) show limited positive correlation with faecal bacteria. Reagent contaminants mainly associated with the Qiagen (light blue) or the Mpbio (green) kit form distinct clusters. Species that are strongly associated with sample collection contamination in 2012–2013 are indicated in orange. For each species the highest value (percentage) found using either the Qiagen or the Mpbio DNA isolation kit, was used as input (using all 498 samples).
Extended Data Fig. 9 Bacterial signals and adverse pregnancy outcome.
a–d, Scatterplot representations of the non-significant associations of S. agalactiae with SGA (a), S. anginosus with SGA (b), and of the significant associations of L. iners with pre-eclampsia (c), and Ureaplasma with PTB (d). Samples with 0% signal are not shown on the logarithmic scale. Signals above 1% (dotted line) are regarded as positive for use in McNemar’s test (a–c), and signals below 1% are considered as negative. The Mann–Whitney U-test was used for unpaired samples in d.
Supplementary information
Supplementary Information
This supplementary information file contains all six Supplementary Tables. It furthermore contains a Supplementary Discussion section and a Supplementary Methods section.
Supplementary Information
This supplementary information file contains detailed oligotype abundance information (16S rRNA amplicon sequencing results).
Rights and permissions
About this article
Cite this article
de Goffau, M.C., Lager, S., Sovio, U. et al. Human placenta has no microbiome but can contain potential pathogens. Nature 572, 329–334 (2019). https://doi.org/10.1038/s41586-019-1451-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1451-5
This article is cited by
-
Early-life ruminal microbiome-derived indole-3-carboxaldehyde and prostaglandin D2 are effective promoters of rumen development
Genome Biology (2024)
-
Placental TLR recognition of salivary and subgingival microbiota is associated with pregnancy complications
Microbiome (2024)
-
A genome-wide association study reveals the relationship between human genetic variation and the nasal microbiome
Communications Biology (2024)
-
Gestational diabetes augments group B Streptococcus infection by disrupting maternal immunity and the vaginal microbiota
Nature Communications (2024)
-
Maternal microbiota communicates with the fetus through microbiota-derived extracellular vesicles
Microbiome (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.