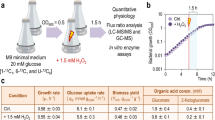
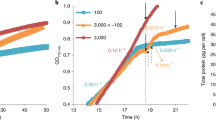
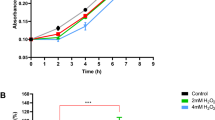
Abstract
Both single and multicellular organisms depend on anti-stress mechanisms that enable them to deal with sudden changes in the environment, including exposure to heat and oxidants. Central to the stress response are dynamic changes in metabolism, such as the transition from the glycolysis to the pentose phosphate pathway—a conserved first-line response to oxidative insults1,2. Here we report a second metabolic adaptation that protects microbial cells in stress situations. The role of the yeast polyamine transporter Tpo1p3,4,5 in maintaining oxidant resistance is unknown6. However, a proteomic time-course experiment suggests a link to lysine metabolism. We reveal a connection between polyamine and lysine metabolism during stress situations, in the form of a promiscuous enzymatic reaction in which the first enzyme of the polyamine pathway, Spe1p, decarboxylates lysine and forms an alternative polyamine, cadaverine. The reaction proceeds in the presence of extracellular lysine, which is taken up by cells to reach concentrations up to one hundred times higher than those required for growth. Such extensive harvest is not observed for the other amino acids, is dependent on the polyamine pathway and triggers a reprogramming of redox metabolism. As a result, NADPH—which would otherwise be required for lysine biosynthesis—is channelled into glutathione metabolism, leading to a large increase in glutathione concentrations, lower levels of reactive oxygen species and increased oxidant tolerance. Our results show that nutrient uptake occurs not only to enable cell growth, but when the nutrient availability is favourable it also enables cells to reconfigure their metabolism to preventatively mount stress protection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Proteomic data are accessible through ProteomeXchange (PXD013373). The kinetic data is available in Table 1, and the raw data used to generate the figures is provided in the Supplementary Tables.
References
Ralser, M. et al. Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress. J. Biol. 6, 10 (2007).
Kuehne, A. et al. Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 59, 359–371 (2015).
Mima, S. et al. Identification of the TPO1 gene in yeast, and its human orthologue TETRAN, which cause resistance to NSAIDs. FEBS Lett. 581, 1457–1463 (2007).
Albertsen, M., Bellahn, I., Krämer, R. & Waffenschmidt, S. Localization and function of the yeast multidrug transporter Tpo1p. J. Biol. Chem. 278, 12820–12825 (2003).
Uemura, T., Tachihara, K., Tomitori, H., Kashiwagi, K. & Igarashi, K. Characteristics of the polyamine transporter TPO1 and regulation of its activity and cellular localization by phosphorylation. J. Biol. Chem. 280, 9646–9652 (2005).
Krüger, A. et al. Tpo1-mediated spermine and spermidine export controls cell cycle delay and times antioxidant protein expression during the oxidative stress response. EMBO Rep. 14, 1113–1119 (2013).
Ludwig, C. et al. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 14, e8126 (2018).
Tomar, P. C., Lakra, N. & Mishra, S. N. Cadaverine. Plant Signal Behav. 8, 10 (2013).
Yamamoto, Y., Miwa, Y., Miyoshi, K., Furuyama, J. & Ohmori, H. The Escherichia coli ldcC gene encodes another lysine decarboxylase, probably a constitutive enzyme. Genes Genet. Syst. 72, 167–172 (1997).
Igarashi, K. et al. Formation of a compensatory polyamine by Escherichia coli polyamine-requiring mutants during growth in the absence of polyamines. J. Bacteriol. 166, 128–134 (1986).
Whitney, P. A. & Morris, D. R. Polyamine auxotrophs of Saccharomyces cerevisiae. J. Bacteriol. 134, 214–220 (1978).
Taxis, C. A safety catch for ornithine decarboxylase degradation. Microb. Cell 2, 174–177 (2015).
Tyagi, A. K., Tabor, C. W. & Tabor, H. Ornithine decarboxylase from Saccharomyces cerevisiae. Purification, properties, and regulation of activity. J. Biol. Chem. 256, 12156–12163 (1981).
Dufe, V. T. et al. A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane. Biochem. J. 405, 261–268 (2007).
Mülleder, M. et al. Functional metabolomics describes the yeast biosynthetic regulome. Cell 167, 553–565 (2016).
Park, J. O. et al. Metabolite concentrations, fluxes and free energies imply efficient enzyme usage. Nat. Chem. Biol. 12, 482–489 (2016).
Bianchi, F. et al. Asymmetry in inward- and outward-affinity constant of transport explain unidirectional lysine flux in Saccharomyces cerevisiae. Sci. Rep. 6, 31443 (2016).
Ough, C. S., Huang, Z., An, D. & Stevens, D. Amino acid uptake by four commercial yeasts at two different temperatures of growth and fermentation: effects on urea excretion and reabsorption. Am. J. Enol. Vitic. 42, 26–40 (1991).
Tucci, A. F. Feedback inhibition of lysine biosynthesis in yeast. J. Bacteriol. 99, 624–625 (1969).
Feller, A., Ramos, F., Piérard, A. & Dubois, E. In Saccharomyces cerevisae, feedback inhibition of homocitrate synthase isoenzymes by lysine modulates the activation of LYS gene expression by Lys14p. Eur. J. Biochem. 261, 163–170 (1999).
Andi, B., West, A. H. & Cook, P. F. Regulatory mechanism of histidine-tagged homocitrate synthase from Saccharomyces cerevisiae. I. Kinetic studies. J. Biol. Chem. 280, 31624–31632 (2005).
Szappanos, B. et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat. Genet. 43, 656–662 (2011).
Stincone, A. et al. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol. Rev. Camb. Philos. Soc. 90, 927–963 (2015).
Nogae, I. & Johnston, M. Isolation and characterization of the ZWF1 gene of Saccharomyces cerevisiae, encoding glucose-6-phosphate dehydrogenase. Gene 96, 161–169 (1990).
Slekar, K. H., Kosman, D. J. & Culotta, V. C. The yeast copper/zinc superoxide dismutase and the pentose phosphate pathway play overlapping roles in oxidative stress protection. J. Biol. Chem. 271, 28831–28836 (1996).
Zhang, J. et al. Engineering an NADPH/NADP+ redox biosensor in yeast. ACS Synth. Biol. 5, 1546–1556 (2016).
Toroser, D., Yarian, C. S., Orr, W. C. & Sohal, R. S. Mechanisms of γ-glutamylcysteine ligase regulation. Biochim. Biophys. Acta Gen. Subj. 1760, 233–244 (2006).
Shenton, D. & Grant, C. M. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 374, 513–519 (2003).
Grüning, N.-M. et al. Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab. 14, 415–427 (2011).
Campbell, K., Vowinckel, J., Keller, M. A. & Ralser, M. Methionine metabolism alters oxidative stress resistance via the pentose phosphate pathway. Antioxid. Redox Signal. 24, 543–547 (2016).
Peter, J. et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 556, 339–344 (2018).
Dever, T. E. & Ivanov, I. P. Roles of polyamines in translation. J. Biol. Chem. 293, 18719–18729 (2018).
Morano, K. A., Grant, C. M. & Moye-Rowley, W. S. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190, 1157–1195 (2012).
Brachmann, C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998).
Mülleder, M., Campbell, K., Matsarskaia, O., Eckerstorfer, F. & Ralser, M. Saccharomyces cerevisiae single-copy plasmids for auxotrophy compensation, multiple marker selection, and for designing metabolically cooperating communities. F1000 Res. 5, 2351 (2016).
Ralser, M. et al. A catabolic block does not sufficiently explain how 2-deoxy-d-glucose inhibits cell growth. Proc. Natl Acad. Sci. USA 105, 17807–17811 (2008).
Campbell, K. et al. Self-establishing communities enable cooperative metabolite exchange in a eukaryote. eLife 4, e09943 (2015).
Kahm, M. et al. grofit: fitting biological growth curves with R. J. Stat. Softw. 33, 1–21 (2010).
Reyes-Becerril, M., Esteban, M. Á., Tovar-Ramírez, D. & Ascencio-Valle, F. Polyamine determination in different strains of the yeast Debaryomyces hansenii by high pressure liquid chromatography. Food Chem. 127, 1862–1865 (2011).
Sasidharan, K., Soga, T., Tomita, M. & Murray, D. B. A yeast metabolite extraction protocol optimised for time-series analyses. PLoS ONE 7, e44283 (2012).
Mo, M. L., Palsson, B. O. & Herrgård, M. J. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst. Biol. 3, 37 (2009).
Heirendt, L. et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v3.0. Nat. Protoc. 14, 639–702 (2019).
Godon, C. et al. The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273, 22480–22489 (1998).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566 (2013).
Acknowledgements
This work was supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001134), the UK Medical Research Council (FC001134) and the Wellcome Trust (FC001134), as well as specific project funding from the Wellcome Trust (IA 200829/Z/16/Z to M.R.) and the ERC (StG260809 to M.R.). V. O.-S. was funded by the Consejo Nacional de Ciencia y Tecnología Mexico (postdoctoral fellowship 232510.), M.T.A. by the Warwick Medical School and D.A.P.N. by the Austrian Science Fund (Doctoral Program Biomolecular Technology of Proteins, FWF W1224). We thank A. Flint, H. Lee and D. Panneman for help with experiments; and A. Carter and E. Morales-Rios (MRC-LMB, Cambridge), M. K. Jensen (DTU), N. Typas and A. Koumoutsi (EMBL Heidelberg), G. Liti (University of Nice) and J. Hallin (Universite Laval) for providing strains, plasmids and enabling us to access their facilities.
Peer review information
Nature thanks Kazuei Igarashi, Jens Nielsen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
V.O.-S. identified cadaverine and the enzyme responsible for lysine decarboxylation, cloned SPE1 and carried out the expression, purification and kinetic characterization of recombinant Spe1p, the quantification of lysine in wild-type and ∆tpo1 strains, the lysine uptake rate experiment in auxotroph and prototroph strains, the growth curve experiment of ∆zwf1 in media supplemented with lysine and methionine, the quantification of GSH–N-ethylmaleimide and polyamines and several experiments to identify the function of cadaverine. J.S.L.Y. performed the H2O2 stress response experiments in yeast exposed to d/l-lysine and, together with D.A.P.N., sourced the materials and performed the NADPH sensor experiments. L.M.-F. performed experiments to uncover a function for cadaverine, cloned ldcC, performed the stress tests with Δzwf1 and analysed ROS levels by flow cytometry. M.T.A. performed metabolic modelling and FBA analysis. S.K. conducted the experiment that assessed the extent of harvesting for all amino acids and H2O2 resistance in wild yeasts; C.C.-M. tested interactions between H2O2 with lysine and methionine; R.H. addressed the effect of cadaverine on growth; J.S. addressed the role of lysine harvesting in mammalian cells; D.A.P.N. assessed H2O2 resistance in P. pastoris; L.H.-D. conducted bacterial stress test experiments; O.M.-L. performed the molecular docking experiments; J.V. conducted proteomic data analysis; M.M. developed the metabolomics method and conducted LC–MS measurements; and M.R. conceived and supervised the study and wrote the first draft of the paper. All authors contributed to the experimental planning and writing up of the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Proteomic analysis.
A proteomic time-course experiment recorded by SWATH-MS6 was re-analysed with a new spectral library and a recent version of Spectronaut (Biognosys) software, increasing the depth of the proteomic analysis. a, Principal component analysis shows that proteome profiles of H2O2-exposed Δtpo1 yeast6 are more different from the wild type than that of a Tpo1p-overexpressing strain and that there is a metabolic adaptation in the lysine pathway. Numbers indicate time points (in minutes) after the oxidative insult. b, Gene Ontology (GO) enrichment analysis shows that a number of biosynthetic processes are significantly enriched in proteins differentially expressed between wild-type and Δtpo1 yeast upon exposure to H2O2. Most of the GO terms that are significantly enriched belong to processes that are typically seen to be activated upon treatment with H2O2, including oxidation–reduction processes, cell cycle, nucleotide synthesis and ribosome43. Metabolic processes affected include carbohydrate- and amino acid metabolism, including two GO terms specific for the lysine pathway. Significance of enrichment over base frequency was calculated using a binomial test44, and P values (red line) were corrected for false discovery rate using Bonferroni (blue) or Benjamini–Hochberg (green) correction. n = 5.
Extended Data Fig. 2 The S. cerevisiae lysine biosynthesis pathway via aminoadipate.
Lys20p/Lys21p, homocitrate synthase; Aco2p, aconitase; Lys4p, homoaconitase; Lys12p, homoisocitrate dehydrogenase; Aro8p, 2-aminoadipate transaminase; Lys2p, α-aminoadipate reductase; Lys9p, saccharopine dehydrogenase (NADP+ and l-glutamate-forming); Lys1p, saccharopine dehydrogenase (NAD+ and l-lysine-forming).
Extended Data Fig. 3 Effect of lysine supplementation on the intracellular concentration of the canonical polyamines and growth.
a, Polyamine content in yeast cells in the mid-log phase grown in SM media with and without 25 μg ml−1 lysine, as quantified by LC–MS/MS using selected reaction monitoring. Bar charts represent the mean ± s.d.; n = 3 biologically independent samples for non-supplemented conditions and n = 4 for supplemented conditions. Unpaired two-tailed Student’s t-test, wild-type cells supplemented versus non-supplemented with lysine: ****P ≤ 0.0001, **P = 0.0066. b, Growth curves of wild-type cells grown in SM media with or without lysine (25 μg ml−1) supplementation. The data represent mean ± s.d.; n = 3 biologically independent samples.
Extended Data Fig. 4 Predictions of the growth-optimized (naive) model for flux balance analysis of lysine harvesting.
We used a revised version of the S. cerevisiae genome-scale metabolic model (iMM904_NADcorrected22). Six additional reactions related to cadaverine biosynthesis were added: two metabolic reactions for ornithine decarboxylase (H + lys_L → CO2 + cadaverine) and spermine synthase (S-adenosyl 3-(methylthio)propylamine + cadaverine → S-methyl-5′-thioadenosine + aminopropyl-cadaverine), and four reactions for transporting and exchanging cadaverine and aminopropylcadaverine. In silico synthetic minimal media constraints were used according to the original model (iMM90441). For excess lysine uptake, the constraint of the l-lysine exchange reaction was fixed to a value of 1. Model simulation (FBA) was performed using the COBRA toolbox for maximum growth in both media conditions.
Extended Data Fig. 5 Lysine and methionine supplementation have no growth-relevant effect on the oxidative capacity of media containing H2O2.
a, H2O2 levels measured by Amplex Red fluorescence in water (left) or SM medium (right) supplemented with l-lysine (25 μg ml−1), d-lysine (25 μg ml−1) or l-methionine (40 μg ml−1) with or without 30 min pre-incubation with 2.5 mM H2O2. The data represent mean ± s.d.; n = 3 independent experiments. b, Growth curves of cells pre-cultured in SM, SM + lysine (SM + Lys, 25 μg ml−1) or SM + methionine (SM + Met, 40 μg ml−1). At mid-log phase (OD600nm ≈ 0.5) cells were pre-incubated for 0 or 30 min with 2.5 mM of H2O2 in the respective pre-culture medium. The data represent mean ± s.d.; n = 3 biologically independent samples.
Extended Data Fig. 6 Effect of lysine supplementation on mammalian cell lines that are auxotrophic for lysine.
a, Growth curves showing the effect of different concentrations of lysine (0.01–10 mM) on mammalian cell growth. Percentage confluence was calculated using the IncuCyte imaging system (Essen Bioscience). The data represent mean ± s.d.; n = 3 biologically independent samples. b, Intracellular lysine concentrations in different mammalian cell lines grown in DMEM with a range of concentrations of lysine (0.01–10 mM). Cells were seeded at 0.5 × 106 cells per 6-cm plate and grown for 48 h before collection. The data represent mean ± s.d.; n = 3 biologically independent samples.
Extended Data Fig. 7 Lysine harvesting protects against H2O2 in different yeast and bacteria species.
a, Effect of the supplementation of lysine on the resistance of mammalian cells to H2O2. Cell biomass was quantified by crystal-violet staining following 24 h treatment with H2O2 at a range of concentrations in cultures supplemented with 0.1–10 mM lysine. The data represent mean ± s.d.; n = 3 biologically independent samples. b, Growth curves of C. tropicalis, the S. cerevisiae laboratory strain BY4741 with repaired auxotrophic loci and two non-laboratory isolates of S. cerevisiae in SM media with or without 25 μg ml−1 lysine. When challenged with 1.5 mM H2O2, cultures supplemented with lysine (pre-cultures were also supplemented) can survive and eventually grow (red versus green curves). c, Spot test of the resistance of P. pastoris to H2O2. Overnight cultures were grown in SM media (potassium phosphate-buffered, pH 6.0) with or without lysine (76 mg l−1) supplementation and were diluted to OD600nm = 1 in water, serial diluted and spotted onto SM media (potassium phosphate-buffered, pH 6.0) with or without lysine supplementation and H2O2. The experiment was repeated with similar results. d, Growth curves of B. subtilis 168 trpC2 when challenged with 1.5 mM H2O2 versus a water control, at mid-log phase in S7 minimal media with or without lysine (40 µg ml−1) supplementation.
Extended Data Fig. 8 Yeast can accumulate high levels of cadaverine and aminopropylcadaverine that are not toxic.
a, b, Cadaverine (a) and aminopropylcadaverine (APC) (b) concentrations in S. cerevisiae wild-type strain overexpressing E. coli LdcC (WT LDC) or carrying the empty plasmid (WT pYX212). Polyamines were derivatized with dansyl chloride and quantified by LC–MS/MS. The data represent mean ± s.d.; n = 3 biologically independent samples. Unpaired two-tailed Student’s t-test: wild-type pYX212 versus WT LDC in non-supplemented conditions ***P = 0.0003 and supplemented conditions ***P = 0.0007 for a; and **P = 0.0054 and ***P = 0.0007 for non-supplemented and supplemented conditions for b. c, Growth curves showing the effect of lysine (250 µg ml−1) on growth when the wild-type strain is overexpressing E. coli LdcC. The data represent mean ± s.d.; n = 3 biologically independent samples.
Extended Data Fig. 9 Cadaverine can partially substitute for canonical polyamines, but only at non-physiologically high concentrations and at a certain pH, and does not protect S. cerevisiae against H2O2 stress.
a, b, Growth curves of Δspe1 strain depleted from spermidine grown in SM media alone or supplemented with 250 mM cadaverine in the absence (a) or presence (b) of 0.1 mM spermidine. The data represent mean ± s.d.; n = 3 biologically independent samples. c, Growth curves showing the effect of lysine supplementation on wild-type and Δspe1 strains carrying a control plasmid (empty) or overexpressing E. coli LdcC (LDC), when grown in SM media buffered or not buffered at pH 5.0. The data represent mean ± s.d.; n = 4 biologically independent samples per experiment. The experiment was repeated twice. d, H2O2 tolerances were determined as described above, but substituting lysine with 5 mM cadaverine. The data represent mean ± s.d.; n = 4 independent experiments. Even in the presence of this high level of cadaverine, the yeast cells do not tolerate higher levels of H2O2.
Supplementary information
41586_2019_1442_MOESM1_ESM.pdf
Supplementary Information Extended Note 1: Cadaverine formation is not toxic to yeast cells. Describes experiments showing that cadaverine is well-tolerated by Saccharomyces cerevisiae until extreme levels are synthesized by the transgenic expression of a bacterial lysine carboxylase in yeast. Extended Note 2: Cadaverine can partially substitute for canonical polyamines, but only at non physiological high concentrations and certain pH. Describes experiments showing that the alternative polyamines (cadaverine and its metabolites) are not generally substituting for the caconical polyamines (putrescine, spermine, spermidine), but have overlapping properties, so can compensate for polyamine deficiency under specific conditions.
41586_2019_1442_MOESM3_ESM.pdf
Supplementary Figure Supplementary Figure 1: Gating strategy for NADPH sensor experiments. This figure represents the gating strategy. Describes the gating strategy used in NADPH sensor experiments.
Rights and permissions
About this article
Cite this article
Olin-Sandoval, V., Yu, J.S.L., Miller-Fleming, L. et al. Lysine harvesting is an antioxidant strategy and triggers underground polyamine metabolism. Nature 572, 249–253 (2019). https://doi.org/10.1038/s41586-019-1442-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1442-6
This article is cited by
-
Intestinal metabolites predict treatment resistance of patients with depression and anxiety
Gut Pathogens (2024)
-
A non-carboxylative route for the efficient synthesis of central metabolite malonyl-CoA and its derived products
Nature Catalysis (2024)
-
Metabolic exchanges are ubiquitous in natural microbial communities
Nature Microbiology (2023)
-
Metabolic heterogeneity and cross-feeding within isogenic yeast populations captured by DILAC
Nature Microbiology (2023)
-
Lysine catabolism reprograms tumour immunity through histone crotonylation
Nature (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.