Abstract
Neuromuscular disorders are often caused by heterogeneous mutations in large, structurally complex genes. Targeting compensatory modifier genes could be beneficial to improve disease phenotypes. Here we report a mutation-independent strategy to upregulate the expression of a disease-modifying gene associated with congenital muscular dystrophy type 1A (MDC1A) using the CRISPR activation system in mice. MDC1A is caused by mutations in LAMA2 that lead to nonfunctional laminin-α2, which compromises the stability of muscle fibres and the myelination of peripheral nerves. Transgenic overexpression of Lama1, which encodes a structurally similar protein called laminin-α1, ameliorates muscle wasting and paralysis in mouse models of MDC1A, demonstrating its importance as a compensatory modifier of the disease1. However, postnatal upregulation of Lama1 is hampered by its large size, which exceeds the packaging capacity of vehicles that are clinically relevant for gene therapy. We modulate expression of Lama1 in the dy2j/dy2j mouse model of MDC1A using an adeno-associated virus (AAV9) carrying a catalytically inactive Cas9 (dCas9), VP64 transactivators and single-guide RNAs that target the Lama1 promoter. When pre-symptomatic mice were treated, Lama1 was upregulated in skeletal muscles and peripheral nerves, which prevented muscle fibrosis and paralysis. However, for many disorders it is important to investigate the therapeutic window and reversibility of symptoms. In muscular dystrophies, it has been hypothesized that fibrotic changes in skeletal muscle are irreversible. However, we show that dystrophic features and disease progression were improved and reversed when the treatment was initiated in symptomatic dy2j/dy2j mice with apparent hindlimb paralysis and muscle fibrosis. Collectively, our data demonstrate the feasibility and therapeutic benefit of CRISPR–dCas9-mediated upregulation of Lama1, which may enable mutation-independent treatment for all patients with MDC1A. This approach has a broad applicability to a variety of disease-modifying genes and could serve as a therapeutic strategy for many inherited and acquired diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The authors declare that the main data supporting the findings of this study are available within the paper and its Supplementary Information.
References
Gawlik, K., Miyagoe-Suzuki, Y., Ekblom, P., Takeda, S. & Durbeej, M. Laminin α1 chain reduces muscular dystrophy in laminin α2 chain deficient mice. Hum. Mol. Genet. 13, 1775–1784 (2004).
Kemaladewi, D. U. et al. Correction of a splicing defect in a mouse model of congenital muscular dystrophy type 1A using a homology-directed-repair-independent mechanism. Nat. Med. 23, 984–989 (2017).
Sunada, Y., Bernier, S. M., Utani, A., Yamada, Y. & Campbell, K. P. Identification of a novel mutant transcript of laminin α2 chain gene responsible for muscular dystrophy and dysmyelination in dy 2J mice. Hum. Mol. Genet. 4, 1055–1061 (1995).
Gawlik, K. I., Harandi, V. M., Cheong, R. Y., Petersén, Å. & Durbeej, M. Laminin α1 reduces muscular dystrophy in dy 2J mice. Matrix Biol. 70, 36–49 (2018).
Gawlik, K. I., Li, J. Y., Petersén, A. & Durbeej, M. Laminin α1 chain improves laminin α2 chain deficient peripheral neuropathy. Hum. Mol. Genet. 15, 2690–2700 (2006).
Maeder, M. L. et al. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013).
Perez-Pinera, P. et al. RNA-guided gene activation by CRISPR–Cas9-based transcription factors. Nat. Methods 10, 973–976 (2013).
Wojtal, D. et al. Spell checking nature: versatility of CRISPR/Cas9 for developing treatments for inherited disorders. Am. J. Hum. Genet. 98, 90–101 (2016).
Ran, F. A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Kemaladewi, D. U., Benjamin, J. S., Hyatt, E., Ivakine, E. A. & Cohn, R. D. Increased polyamines as protective disease modifiers in congenital muscular dystrophy. Hum. Mol. Genet. 27, 1905–1912 (2018).
Bönnemann, C. G. et al. Diagnostic approach to the congenital muscular dystrophies. Neuromuscul. Disord. 24, 289–311 (2014).
Homma, S., Beermann, M. L. & Miller, J. B. Peripheral nerve pathology, including aberrant Schwann cell differentiation, is ameliorated by doxycycline in a laminin-α2-deficient mouse model of congenital muscular dystrophy. Hum. Mol. Genet. 20, 2662–2672 (2011).
Qiao, C. et al. Amelioration of muscle and nerve pathology in LAMA2 muscular dystrophy by AAV9-mini-agrin. Mol. Ther. Methods Clin. Dev. 9, 47–56 (2018).
Patton, B. L., Wang, B., Tarumi, Y. S., Seburn, K. L. & Burgess, R. W. A single point mutation in the LN domain of LAMA2 causes muscular dystrophy and peripheral amyelination. J. Cell Sci. 121, 1593–1604 (2008).
Previtali, S. C. et al. Expression of laminin receptors in Schwann cell differentiation: evidence for distinct roles. J. Neurosci. 23, 5520–5530 (2003).
Bentzinger, C. F., Barzaghi, P., Lin, S. & Ruegg, M. A. Overexpression of mini-agrin in skeletal muscle increases muscle integrity and regenerative capacity in laminin-α2-deficient mice. FASEB J. 19, 934–942 (2005).
Reinhard, J. R. et al. Linker proteins restore basement membrane and correct LAMA2-related muscular dystrophy in mice. Sci. Transl. Med. 9, eaal4649 (2017).
McKee, K. K. et al. Chimeric protein repair of laminin polymerization ameliorates muscular dystrophy phenotype. J. Clin. Invest. 127, 1075–1089 (2017).
Rooney, J. E., Gurpur, P. B. & Burkin, D. J. Laminin-111 protein therapy prevents muscle disease in the mdx mouse model for Duchenne muscular dystrophy. Proc. Natl Acad. Sci. USA 106, 7991–7996 (2009).
Perrin, A., Rousseau, J. & Tremblay, J. P. Increased expression of laminin subunit alpha 1 chain by dCas9–VP160. Mol. Ther. Nucleic Acids 6, 68–79 (2017).
Yuan, J. et al. Genetic modulation of RNA splicing with a CRISPR-guided cytidine deaminase. Mol Cell 72, 380–394 (2018).
Villiger, L. et al. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 24, 1519–1525 (2018).
Bengtsson, N. E. et al. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 8, 14454 (2017).
Liao, H. K. et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell 171, 1495–1507 (2017).
Thakore, P. I. et al. Highly specific epigenome editing by CRISPR–Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149 (2015).
Zhang, Y. et al. Notch1 signaling plays a role in regulating precursor differentiation during CNS remyelination. Proc. Natl Acad. Sci. USA 106, 19162–19167 (2009).
Hakim, C. H. et al. A five-repeat micro-dystrophin gene ameliorated dystrophic phenotype in the severe DBA/2J-mdx model of Duchenne muscular dystrophy. Mol. Ther. Methods Clin. Dev. 6, 216–230 (2017).
Acknowledgements
We thank members of the Cohn laboratory for technical support and input in this study; P. Yurchenco for providing reagents used in this study, C. Rand, I. Vukobradovic and R. Smith for assistance with functional and behavioural studies, D. Holmyard for transmission electron microscopy analysis and S. Pereira for genomic data acquisition. This work was supported by AFM-Telethon, Cure CMD, Muscular Dystrophy Association (to D.U.K.), Rare Disease Foundation Microgrant (to P.S.B. and S.E.), SickKids Restracomp (to S.E.), Crafoord Foundation (to K.I.G.), CIHR Summer Studentship (to R.M.M.), Swedish Research Council, Anna and Edwin Berger Foundation, Greta and Johan Kock Foundation, Olle Engkvist Byggmastare Foundation and Osterlund Foundation (to M.D.), Natural Sciences and Engineering Research Council of Canada, Canadian Institute for Health Research, SickKids Foundation and R. S. McLaughlin Foundation Chair in Pediatrics (to R.D.C.).
Reviewer information
Nature thanks James M. Ervasti, Reid D. Landes, Daniel Michele and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
D.U.K., E.A.I. and R.D.C. conceived and supervised the study. P.S.B., K.L., R.K. and K.M.P. cloned and screened guides in vitro. D.U.K., P.S.B., K.L. and E.H. performed in vivo injections and contractile assays. D.U.K., P.S.B., K.L., K.M.P. and R.M.M. analysed muscle histology. K.I.G. and M.D. performed extended muscle imaging and provided reagents. D.U.K. performed longitudinal open field assay and vector genome quantification. D.U.K. and P.S.B. performed nerve imaging with the help from K.M.P. and R.M.M. D.U.K. and D.A.-B. performed nerve conduction velocity under supervision by S.A.P. S.E. analysed RNA-sequencing data and off-target prediction. D.U.K. wrote the manuscript with inputs from the other authors. All authors provided feedback and agreed on the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 SadCas9–2×VP64 enhances expression of minCMV-driven tdTomato in vitro.
a, b, HEK293T cells were transfected with a plasmid encoding minCMV-driven tdTomato only. c, d, HEK293T cells were transfected with a combination of a plasmid containing minCMV-driven tdTomato and one containing SadCas9–2×VP64 and a sgRNA targeting the minCMV promoter. a, c, Schematic. b, d, Cells were imaged for tdTomato expression by fluorescent microscopy. Bright-field and tdTomato images are shown. The experiment was repeated three times independently with similar results. Scale bars, 50 µm.
Extended Data Fig. 2 Early intervention to upregulate Lama1 prevents disease progression in dy2j/dy2j mice.
a, P2 neonatal dy2j/dy2j mice were injected with AAV9s carrying no guide (n = 6; 7.5 × 1011 viral genomes) or three guides (n = 4; split into two vectors; total dose of 2 × 7.5 × 1011 viral genomes) in the temporal vein and euthanized 7 weeks later. b, c, Immunofluorescence staining shows Lama1 expression in tibialis anterior (b) and gastrocnemius (c) muscles. Scale bars, 100 µm. d, e, General muscle histopathology was evaluated by haematoxylin and eosin staining on tibialis anterior (d) and gastrocnemius (e) muscles. Scale bars, 200 µm. All mice (n = 6, no guide and n = 4, three guides) were analysed for experiments shown in b–e, which were repeated three times independently with similar results. f–i, Fibrosis (f, h) and minimum Feret diameter (g, i) were quantified for all mice from both treatment groups (n = 6, no guide and n = 4, three guides) by two independent experimenters with similar results. Data are mean ± s.d. **P = 0.0025 (f), *P = 0.0103 (g), *P = 0.0142 (h), P = 0.0711 (i); two-tailed Student’s t-test.
Extended Data Fig. 3 Upregulation of Lama1 corresponds to improvement of muscle functions.
Three-week-old dy2j/dy2j mice were injected systemically in the tail vein with AAV9s carrying no guide (n = 9) at the dose of 3 × 1011 viral genomes (vg) per gram of mouse, or three guides at different doses of 7.5 × 1010 (n = 4), 1.5 × 1011 (n = 3) or 3 × 1011 (n = 8) viral genome per gram of mouse. Two AAVs were needed for the three-guide cohorts, therefore the total doses of virus injected were 1.5 × 1011, 3 × 1011 and 6 × 1011 viral genomes per gram of mouse. a, Tibialis anterior muscles isolated that were isolated four weeks later were stained for Lama1 expression. Asterisks indicate Lama1-positive fibres in the low-dose cohort. The experiment was repeated three times independently with similar results. Scale bars, 100 µm. b, c, In vivo contractile force assays (b) and open field tests (c) were performed at the end of the treatment regimen. Data are mean ± s.d. b, P values from left to right are: P = 0.9849, P = 0.9082, **P = 0.0044; one-way ANOVA with Dunnett’s multiple comparisons test. c, P values from left to right are: P = 0.9640, P = 0.9646, **P = 0.0085; one-way ANOVA with Dunnett’s multiple comparisons test.
Extended Data Fig. 4 Representative images of Lama1-positive muscle sections.
Three-week-old mice were injected systemically with three guides (n = 5) at the dose of 3 × 1011 viral genomes per gram of mouse (split into two vectors, thus total dose was 2 × 3× 1011 viral genomes per gram of mouse) in the tail vein, and euthanized at the age of 11–12 weeks old. Muscles were stained for Lama1 expression (red). Scale bars, 500 µm. The experiment was repeated three times independently with similar results.
Extended Data Fig. 5 Expression of laminin subunits in tibialis anterior muscles.
Expression of laminin (LM) subunits in tibialis anterior muscles of wild-type, dy2j/dy2j (intravenously injected with AAVs carrying no guide at 3 × 1011 viral genomes per gram of mouse or three guides at a total dose of 2 × 3× 1011 viral genomes per gram of mouse) and dy2j/dy2j LMα1 (with transgenic overexpression of LMα1) mice (n = 3 for each group). Expression of the LMα1 chain in the tibialis anterior muscle is comparable between dy2j/dy2j mice treated with three guides and transgenic mice. No major differences in expression of LMα2 and LMγ1 were detected between the groups. The LMα4 chain is upregulated in muscles of dy2j/dy2j mice treated with the no-guide AAV and the expression of the LMα4 chain is partially normalized in muscle of dy2j/dy2j mice treated with the three-guide AAV. Scale bar, 50 μm. The experiment was repeated three times independently with similar results.
Extended Data Fig. 6 Expression of laminin subunits in gastrocnemius muscles.
Expression of laminin subunits in gastrocnemius muscles from wild-type, dy2j/dy2j (intravenously injected with AAVs carrying no guide at 3 × 1011 viral genomes per gram of mouse or three guides at a total dose of 2 × 3 × 1011 viral genomes per gram of mouse) and dy2j/dy2j LMα1 (with transgenic overexpression of LMα1) mice (n = 3 for each group). Expression of the LMα1 chain in the gastrocnemius muscles is comparable between dy2j/dy2j mice treated with three guides and transgenic mice. No major differences in expression of LMα2 and LMγ1 were detected between the groups. The LMα4 chain is upregulated in muscle of dy2j/dy2j mice treated with the no-guide AAV and the expression of the LMα4 chain is partially normalized in muscle of dy2j/dy2j AAV-mice treated with three guides. Scale bar, 50 μm. The experiment was repeated three times independently with similar results.
Extended Data Fig. 7 Myelination of sciatic nerves.
Transmission electron microscopy images of sciatic nerves isolated from dy2j/dy2j mice injected with AAV9s carrying no guide (3 × 1011 viral genomes per gram of mouse) or three guides (split into two vectors; total dose of 2 × 3 × 1011 viral genomes per gram of mouse). Three mice were analysed per group. Myelin thickness was measured as the difference between the distance of the myelin and axon (m+a, blue solid line) and axon only (a; red dotted line), quantified using ImageJ and presented as mean ± s.d. Statistical analysis was performed using a two-tailed t-test. *P = 0.0231. Scale bar, 5 μm. The experiment was repeated three times independently with similar results.
Extended Data Fig. 8 Quantitative evaluation of AAV genome distribution.
Genomic DNA isolated from the tibialis anterior muscle, sciatic nerve and liver of dy2j/dy2j mice (n = 8) injected with AAVs carrying three guides at the dose of 3 × 1011 viral genomes per gram of mouse (split into two vectors; total dose of 2 × 3 × 1011 viral genomes per gram of mouse) was amplified to analyse the presence of viral genomes by qPCR. Data are mean ± s.e.m. The experiment was repeated three times independently with similar results.
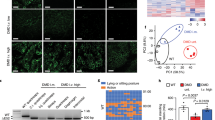
Extended Data Fig. 9 Genome-wide analysis of gene expression of SadCas9-2×VP64-treated dy2j/dy2j mice.
a–c, Differential expression analysis derived from RNA-sequencing results from the quadriceps of treated and untreated mice (n = 3 per group). a, b, No guide: dy2j/dy2j mice injected with AAV9 carrying only the SadCas9–2×VP64 at the dose of 3 × 1011 viral genomes per gram of mouse. a, c, Three guides: dy2j/dy2j mice injected with AAV9 carrying the SadCas9–2×VP64 and three guides at the dose of 3 × 1011 viral genomes per gram of mouse (total dose of 2 × 3 × 1011 viral genomes per gram of mouse). b, c, Untreated: dy2j/dy2j mice that had not been injected with AAV9. False discovery and multiple testing were controlled for and an adjusted P value was computed using the Benjamini–Hochberg method. Differentially expressed genes were defined as genes with a more than twofold change and adjusted P < 0.05. For each comparison, significantly differentially expressed genes (false-discovery rate-adjusted P < 0.05) are coloured. Red data points indicate a log-transformed fold change in expression greater than one; blue data points indicate a log-transformed fold change in expression less than one. d, Hierarchical clustering was performed on the normalized counts-per-million expression data.
Extended Data Fig. 10 Top 500 differentially expressed genes.
Heat map illustrating the log-normalized counts per million for the top 500 genes that were differentially expressed in quadriceps isolated from wild-type mice, or dy2j/dy2j mice injected with AAV9s carrying SadCas9–2×VP64 only (no guide, n = 3) or with three guides (three guides, n = 3), and compared to age-matched, untreated dy2j/dy2j mice. Red indicates higher expression; blue indicates lower expression.
Supplementary information
Supplemental Figure 1
Uncropped scans with size marker indications Boxes indicate area presented in the corresponding manuscript figures.
Supplemental Method 1
Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines Detailed reporting of design, experimental procedures, and experimental animals in this study.
Supplementary Tables
This file contains Supplementary Tables 1-6 (legends included at the top of each sheet).
Supplementary Video 1
Phenotype of control dy2j/dy2j mouse following early intervention. The dy2j/dy2j mouse was injected with AAV9 containing SadCas9-2xVP64 (no guide) at P2 (pre-symptomatic stage) via temporal vein and video was taken at the age of 7-week old. Hind limb paralysis, contracture and kyphosis resulting from lack of functional Lama2 and compensatory Lama1 are apparent.
Supplementary Video 2
Phenotype of treated dy2j/dy2j mouse following early intervention. The dy2j/dy2j mouse was injected with AAV9 containing SadCas9-2xVP64 and sgRNAs targeting Lama1 proximal promoter (three guides) at P2 (pre-symptomatic stage) via temporal vein and video was taken at the age of 7-week old. Upregulation of compensatory Lama1 expression ameliorates the hind limb paralysis, contracture and kyphosis.
Supplementary Video 3
Phenotype of control dy2j/dy2j mouse following intervention at symptomatic stage. The dy2j/dy2j mouse was injected with AAV9 containing SadCas9-2xVP64 (no guide) at 3-week old (pre-symptomatic stage) via tail vein. Video was taken at the age of 7-week old. Hind limb paralysis, contracture and kyphosis resulting from lack of functional Lama2 and compensatory Lama1 are apparent.
Supplementary Video 4
Phenotype of treated dy2j/dy2j mouse following intervention at symptomatic stage. The dy2j/dy2j mouse was injected with AAV9 containing SadCas9-2xVP64 and sgRNAs targeting Lama1 proximal promoter (three guides) at 3-week old (pre-symptomatic stage) via tail vein. Video was taken at the age of 7-week old. Dystrophic features and disease progression were significantly improved and partially reversed following upregulation of Lama1.
Rights and permissions
About this article
Cite this article
Kemaladewi, D.U., Bassi, P.S., Erwood, S. et al. A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature 572, 125–130 (2019). https://doi.org/10.1038/s41586-019-1430-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1430-x
This article is cited by
-
Comparison of DNA targeting CRISPR editors in human cells
Cell & Bioscience (2023)
-
Profiling the impact of the promoters on CRISPR-Cas12a system in human cells
Cellular & Molecular Biology Letters (2023)
-
Advances in CRISPR therapeutics
Nature Reviews Nephrology (2023)
-
Compact engineered human mechanosensitive transactivation modules enable potent and versatile synthetic transcriptional control
Nature Methods (2023)
-
mRNA trans-splicing dual AAV vectors for (epi)genome editing and gene therapy
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.