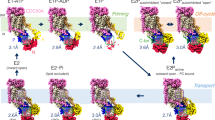
Abstract
Type 4 P-type ATPases (P4-ATPases) are lipid flippases that drive the active transport of phospholipids from exoplasmic or luminal leaflets to cytosolic leaflets of eukaryotic membranes. The molecular architecture of P4-ATPases and the mechanism through which they recognize and transport lipids have remained unknown. Here we describe the cryo-electron microscopy structure of the P4-ATPase Drs2p–Cdc50p, a Saccharomyces cerevisiae lipid flippase that is specific to phosphatidylserine and phosphatidylethanolamine. Drs2p–Cdc50p is autoinhibited by the C-terminal tail of Drs2p, and activated by the lipid phosphatidylinositol-4-phosphate (PtdIns4P or PI4P). We present three structures that represent the complex in an autoinhibited, an intermediate and a fully activated state. The analysis highlights specific features of P4-ATPases and reveals sites of autoinhibition and PI4P-dependent activation. We also observe a putative lipid translocation pathway in this flippase that involves a conserved PISL motif in transmembrane segment 4 and polar residues of transmembrane segments 2 and 5, in particular Lys1018, in the centre of the lipid bilayer.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Cryo-EM maps for the S. cerevisiae Drs2p–Cdc50p in the E2Pinhib, E2Pinter and E2Pactive forms are available on the Electron Microscopy Data Bank under accession numbers EMD-4972 (E2Pinhib; that is, E2–BeF3−), EMD-4973 (E2Pinter; that is, E2–BeF3−–PI4P) and EMD-4974 (E2Pactive; that is, C-terminally truncated E2–BeF3−–PI4P). Coordinates of the atomic structures have been deposited in the PDB under accession numbers 6ROH, 6ROI and 6ROJ.
References
Hankins, H. M., Baldridge, R. D., Xu, P. & Graham, T. R. Role of flippases, scramblases and transfer proteins in phosphatidylserine subcellular distribution. Traffic 16, 35–47 (2015).
Montigny, C., Lyons, J., Champeil, P., Nissen, P. & Lenoir, G. On the molecular mechanism of flippase- and scramblase-mediated phospholipid transport. Biochim. Biophys. Acta 1861, 767–783 (2016).
Pomorski, T. G. & Menon, A. K. Lipid somersaults: uncovering the mechanisms of protein-mediated lipid flipping. Prog. Lipid Res. 64, 69–84 (2016).
Yang, Y., Lee, M. & Fairn, G. D. Phospholipid subcellular localization and dynamics. J. Biol. Chem. 293, 6230–6240 (2018).
Ding, J. et al. Identification and functional expression of four isoforms of ATPase II, the putative aminophospholipid translocase. Effect of isoform variation on the ATPase activity and phospholipid specificity. J. Biol. Chem. 275, 23378–23386 (2000).
Jacquot, A. et al. Phosphatidylserine stimulation of Drs2p·Cdc50p lipid translocase dephosphorylation is controlled by phosphatidylinositol-4-phosphate. J. Biol. Chem. 287, 13249–13261 (2012).
Coleman, J. A., Vestergaard, A. L., Molday, R. S., Vilsen, B. & Andersen, J. P. Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc. Natl Acad. Sci. USA 109, 1449–1454 (2012).
Lee, J. Y. et al. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature 533, 561–564 (2016).
Perez, C. et al. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature 524, 433–438 (2015).
Brunner, J. D., Lim, N. K., Schenck, S., Duerst, A. & Dutzler, R. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 (2014).
Morra, G. et al. Mechanisms of lipid scrambling by the G protein-coupled receptor opsin. Structure 26, 356–367 (2018).
Baldridge, R. D. & Graham, T. R. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc. Natl Acad. Sci. USA 110, E358–E367 (2013).
Jensen, M. S. et al. Phospholipid flipping involves a central cavity in P4 ATPases. Sci. Rep. 7, 17621 (2017).
Vestergaard, A. L. et al. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc. Natl Acad. Sci. USA 111, E1334–E1343 (2014).
Bryde, S. et al. CDC50 proteins are critical components of the human class-1 P4-ATPase transport machinery. J. Biol. Chem. 285, 40562–40572 (2010).
Coleman, J. A. & Molday, R. S. Critical role of the β-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J. Biol. Chem. 286, 17205–17216 (2011).
van der Mark, V. A., Elferink, R. P. & Paulusma, C. C. P4 ATPases: flippases in health and disease. Int. J. Mol. Sci. 14, 7897–7922 (2013).
Alder-Baerens, N., Lisman, Q., Luong, L., Pomorski, T. & Holthuis, J. C. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell 17, 1632–1642 (2006).
Natarajan, P., Wang, J., Hua, Z. & Graham, T. R. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc. Natl Acad. Sci. USA 101, 10614–10619 (2004).
Azouaoui, H. et al. A high-yield co-expression system for the purification of an intact Drs2p–Cdc50p lipid flippase complex, critically dependent on and stabilized by phosphatidylinositol-4-phosphate. PLoS ONE 9, e112176 (2014).
Zhou, X. & Graham, T. R. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl Acad. Sci. USA 106, 16586–16591 (2009).
Xu, P., Baldridge, R. D., Chi, R. J., Burd, C. G. & Graham, T. R. Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J. Cell Biol. 202, 875–886 (2013).
Natarajan, P. et al. Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat. Cell Biol. 11, 1421–1426 (2009).
Zhou, X., Sebastian, T. T. & Graham, T. R. Auto-inhibition of Drs2p, a yeast phospholipid flippase, by its carboxyl-terminal tail. J. Biol. Chem. 288, 31807–31815 (2013).
Tsai, P. C., Hsu, J. W., Liu, Y. W., Chen, K. Y. & Lee, F. J. Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc. Natl Acad. Sci. USA 110, E668–E677 (2013).
Azouaoui, H. et al. High phosphatidylinositol 4-phosphate (PI4P)-dependent ATPase activity for the Drs2p–Cdc50p flippase after removal of its N- and C-terminal extensions. J. Biol. Chem. 292, 7954–7970 (2017).
Puts, C. F. et al. Mapping functional interactions in a heterodimeric phospholipid pump. J. Biol. Chem. 287, 30529–30540 (2012).
Møller, J. V., Olesen, C., Winther, A. M. & Nissen, P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q. Rev. Biophys. 43, 501–566 (2010).
Chantalat, S. et al. The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J. Cell Sci. 117, 711–722 (2004).
Morth, J. P. et al. Crystal structure of the sodium–potassium pump. Nature 450, 1043–1049 (2007).
Poulsen, H. et al. Neurological disease mutations compromise a C-terminal ion pathway in the Na+/K+-ATPase. Nature 467, 99–102 (2010).
Price, E. M. & Lingrel, J. B. Structure–function relationships in the Na,K-ATPase α subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry 27, 8400–8408 (1988).
Obara, K. et al. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+. Proc. Natl Acad. Sci. USA 102, 14489–14496 (2005).
Olsson, M. H., Søndergaard, C. R., Rostkowski, M. & Jensen, J. H. PROPKA3: consistent treatment of internal and surface residues in empirical pK a predictions. J. Chem. Theory Comput. 7, 525–537 (2011).
Jakobi, A. J., Wilmanns, M. & Sachse, C. Model-based local density sharpening of cryo-EM maps. eLife 6, e27131 (2017).
Sørensen, T. L., Møller, J. V. & Nissen, P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science 304, 1672–1675 (2004).
Dolinsky, T. J., Nielsen, J. E., McCammon, J. A. & Baker, N. A. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 (2004).
Jurrus, E. et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018).
Møller, J. V., Lind, K. E. & Andersen, J. P. Enzyme kinetics and substrate stabilization of detergent-solubilized and membraneous (Ca2+ + Mg2+)-activated ATPase from sarcoplasmic reticulum. Effect of protein–protein interactions. J. Biol. Chem. 255, 1912–1920 (1980).
Azouaoui, H. et al. Coordinated overexpression in yeast of a P4-ATPase and its associated Cdc50 subunit: the case of the Drs2p/Cdc50p lipid flippase complex. Methods Mol. Biol. 1377, 37–55 (2016).
Baginski, E. S., Foa, P. P. & Zak, B. Determination of phosphate: study of labile organic phosphate interference. Clin. Chim. Acta 15, 155–158 (1967).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Grant, T. & Grigorieff, N. Automatic estimation and correction of anisotropic magnification distortion in electron microscopes. J. Struct. Biol. 192, 204–208 (2015).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zivanov, J., Nakane, T. & Scheres, S. H. W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019).
Fernandez-Leiro, R. & Scheres, S. H. W. A pipeline approach to single-particle processing in RELION. Acta Crystallogr. D 73, 496–502 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Wang, S., Li, W., Liu, S. & Xu, J. RaptorX-Property: a web server for protein structure property prediction. Nucleic Acids Res. 44, W430–435 (2016).
Kidmose, R. T. et al. Namdinator - automatic Molecular Dynamics flexible fitting of structural models into cryo-EM and crystallography experimental maps. IUCrJ. 6, https://doi.org/10.1107/S2052252519007619 (2019).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Yan, R. et al. Human SEIPIN binds anionic phospholipids. Dev. Cell 47, 248–256 (2018).
Ashkenazy, H. et al. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 44, W344–W350 (2016).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Afonine, P. V. et al. New tools for the analysis and validation of cryo-EM maps and atomic models. Acta Crystallogr. D 74, 814–840 (2018).
Acknowledgements
We thank A. M. Nielsen and T. Klymchuk for technical assistance; P. Gourdon and C. Grønberg for early contributions on samples; P. Champeil for initial functional characterization of purified Drs2p–Cdc50p and for critically reading the manuscript; D. Mills and colleagues at the Max Planck Institute for Biophysics (Frankfurt/Main) and R. Liebrechts at the iNANO center (Aarhus University) for support with data collection and discussions. This work was supported by grants from the Danish National Research Foundation for the PUMPkin center of excellence and from the Lundbeck Foundation for the Brainstruc center of excellence (2015-2666) to P.N.; by an EMBO Long-Term Fellowship to M.-R.A.; by postdoctoral grants from the Danish Council for Independent Research (0602-02912B) and the Lundbeck Foundation (R171-2014-663 and R209-2015-2704) to J.A.L.; by a PhD fellowship from the Boehringer-Ingelheim Fonds to M.T.; by an ANR grant (ANR-14-CE09-0022), the French Infrastructure for Integrated Structural Biology (FRISBI; ANR-10-INSB-05) and the Centre National de la Recherche Scientifique (CNRS) to G.L.; and by the German Research Foundation to A.M. (Mo2752/2).
Peer review information
Nature thanks Joost Holthuis and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
P.N. and G.L. conceived the project, and J.A.L., T.B. and P.N. defined the cryo-EM study with A.M. and W.K. The samples were characterized and developed by M.T., J.J.U., J.A.L. and M.-R.A., together with T.D., C.M. and G.L., and exploratory electron microscopy studies were performed by J.A.L., M.T., J.J.U. and T.B. Cryo-EM analysis was performed by M.T., D.J., J.A.L., T.B. and A.M. Data processing and 3D reconstruction were performed by M.T., with support and advice from D.J., J.A.L., J.L.K. and A.M. Model building and refinement were performed by M.T. and J.A.L., with assistance from J.J.U. and J.L.K. Mutant forms were prepared and functionally characterized by T.D., C.M. and G.L. P.N. and J.A.L. supervised the project together with A.M. The manuscript was drafted by M.T., J.A.L. and P.N. All authors commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Activity of samples used for structural studies.
a, Post–Albers cycle for the Na+, K+-ATPase. Sodium is transported during the E1 half-cycle, and potassium during the E2 half-cycle. Pi, inorganic phosphate. b, Post–Albers cycle for Drs2p–Cdc50p, indicating the off-cycle regulation (grey). Lipid is transported during the E2 half-cycle. The structures that were determined in this work are marked with red circles. PL, translocated phospholipid. c, Topology of Drs2p–Cdc50p, indicating the cleavages at the termini of Drs2p of the constructs that were used for structural studies (ΔN104: all constructs; ΔC1247: E2Pactive). Cdc50p is pink; for Drs2p the transmembrane domain is tan, the A, P and N domains are yellow, blue and red, respectively, and the autoinhibitory C terminus is green. d, Specific activity of Drs2pΔN104/C1247–Cdc50p and Drs2pΔN104–Cdc50p in LMNG, measured by Baginski assay. Water-soluble phosphatidylserine (C8:0), brain PI4P and BeF3− were added as indicated to final concentrations of 78 µg ml−1, 20 µg ml−1 and 5 mM, respectively. Data are mean ± s.d. of four replicates from two independent purification batches. e, Alignment of Drs2p–Cdc50p (in the E2Pactive form) and SERCA (PDB 3B9B) (superpositioning of the Cα carbons, excluding the N domain). Drs2p–Cdc50p is green; SERCA is orange. f, The phosphorylation site of E2Pactive, showing density for the BeF3− inhibitor, Mg2+ ion and coordinating residues. The characteristic E2P conformation of the dephosphorylation loop, in which the glutamate points away from the phosphorylation site, is shown in stick representation. g, The three Drs2p–Cdc50p structures aligned based on the P domain. Asp560–BeF3− and Glu342 are shown to illustrate the similar conformations. Colours are as in Fig. 1a.
Extended Data Fig. 2 Purification and negative-stain electron microscopy of samples for structural studies.
a, SEC chromatogram and gel of Drs2pΔN104–Cdc50p. The red line on the SDS–PAGE image indicates the pooled fractions. b, SEC chromatogram and gel of Drs2pΔN104/C1247–Cdc50p; gel shows the SEC fractions. Red line as in a. c, Analytical SEC of Drs2pΔN104/C1247–Cdc50p purified from b; this was used for cryo-EM. d, A representative negative-stained micrograph of autoinhibited Drs2pΔ104–Cdc50p in LMNG. e, Representative 2D class averages of the sample in d show well-defined and homogeneous particles with recognizable features of P-type ATPases. f, Enlarged 2D class average, highlighting the recognizable domains of Drs2p–Cdc50p.
Extended Data Fig. 3 Fold of Cdc50p.
a, Cdc50p from E2Pinhib, shown in rainbow colours. Cα carbons of glycosylated asparagine residues are shown as black spheres; Drs2p is shown in grey. b, Position of the disulfide sites in Cdc50p from E2Pinhib. c–e, Glycosylation sites in Cdc50p from E2Pinhib. Map levels are 1.75 r.m.s.d. in c and d. The density for the glycosylation at Asn237 is only apparent at lower map levels (0.75 r.m.s.d.) and has not been modelled. f, Alignment of Cdc50p (magenta cartoon) and a monomer of human seipin57, a lipid-binding protein (blue cartoon, PDB 6DS5), illustrating the similar folds (although some loops of Cdc50p are more extensive). The sequence identity between the two proteins is only 4%. Transmembrane helices of seipin that extend from similar positions to the helices of Cdc50p are not present in the structure, but may extend into the membrane in a similar manner to that observed for Cdc50p. g, The transmembrane helices of the Drs2p–Cdc50p complex viewed from the luminal side. The transmembrane helices of Drs2p are rainbow coloured; Cdc50p transmembrane helices are pink. h, Interaction between the Cdc50p ectodomain (shown as surface) and the luminal TM3–TM4 loop of Drs2p (light green). i, Interaction between the N terminus of Cdc50p and the segment of Drs2p that leads from TM4 to the phosphorylation site. Residues 529–538 are not present in P2-ATPases and are shown in dark blue. j, Segments of Drs2p that were found to interact with Cdc50p mutants that disrupt the formation of the complex are highlighted in green, and the insert in Drs2p between TM4 and the phosphorylation site is blue. The structure shown in h–j is E2Pinhib. Cdc50p is coloured on the basis of the conservation using ConSurf58. k, Part of a sequence alignment of proteins of the CDC50 family from S. cerevisiae (Cdc50p, Lem3p and Crf1p) and human CDC50A and CDC50B. Residues that are important for complex formation are identified. Uniprot identifiers are as follows: Cdc50p, P25656; Lem3p, P42838; Crf1p, P53740; CDC50A, Q9NV96; CDC50B, Q3MIR4. The alignment was performed using Clustal Omega59. For full sequence alignment, see Supplementary Fig. 1.
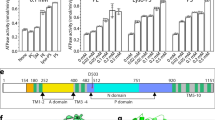
Extended Data Fig. 4 Characterization of PI4P-binding mutants.
a, Coomassie-blue-stained SDS–PAGE of streptavidin-purified wild-type and mutant Drs2p–Cdc50p. Tobacco etch virus protease (TEV) was used to release the complex from the streptavidin beads. b, ATPase activity of PI4P-binding mutants, using an enzyme-coupled assay. The assay medium contained 1 mM ATP, 0.1 mg ml−1 POPS and 1 mg ml−1 DDM in SSR buffer. The rate of ATP hydrolysis was continuously recorded at 340 nm after subsequent addition of 2 µg ml−1 of the purified complex, 5 µg ml−1 trypsin and 25 µg ml−1 PI4P. c, SEC on a Superdex 200 10/300GL column. The arrow indicates the dead volume (V0). d, Limited proteolysis of streptavidin-purified wild-type and mutant Drs2p–Cdc50p by thrombin (Thr.). Control sample in lane 1 contains full-length (FL) Drs2p–Cdc50p. In the absence of PI4P, wild-type and mutant forms of Drs2p are truncated to sizes that correspond to the loss of the first 104 N-terminal residues (ΔN Drs2p; top band in lanes 2, 4, 6 and 8) and to the loss of both the first 104 N-terminal residues and the last 65 C-terminal residues (ΔN/C Drs2p; second band in lanes 2, 4, 6 and 8). Incubation with PI4P during limited proteolysis (lanes 3, 5, 7 and 9) promotes further C-terminal truncation only for the wild-type Drs2p, indicating that the Drs2p mutants are less sensitive to PI4P. e, Control proteolysis experiment showing that phosphatidylserine does not alter the cleavage of wild-type Drs2p–Cdc50p with thrombin, and thereby demonstrating that the C-terminal mutants Y1235A, Y1235F and H1236A are specifically defective in PI4P binding.
Extended Data Fig. 5 Proposed lipid-translocation pathway and comparison between the three Drs2p–Cdc50p structures.
a, A proposed pathway for the translocation of lipids is revealed after activation by PI4P binding and C-terminal truncation. Electrostatic surfaces were calculated in APBS37,38. See also Fig. 4a. b, Binding of potassium in the Na+, K+-ATPase30 (PDB 3KDP), showing the coordination of the ions by negatively charged and polar residues. TM4 is coloured yellow, the PEGL motif is orange and the bound potassium ions are purple spheres. c, Sites and residues of E2Pactive that correspond to the ion-binding sites of the Na+, K+-ATPase in b. TM4 is yellow, the PISL motif is orange and stabilizing hydrogen bonds are shown. d, Superpositioning of the three Drs2p–Cdc50p structures, based on Cdc50p and TM7–TM10 of Drs2p. E2Pinhib is green, E2Pinter is blue and E2Pactive is yellow. The major conformational changes are in the cytosolic domains, TM1 and TM2. e, Expanded view of d, highlighting the changes in the P domains of the three Drs2p–Cdc50p structures. The TM6–TM7 loop and the H1C-tail are depicted in slightly darker colours and the autoinhibitory domain of E2Pinhib is dark green. Destabilization or removal of the helical segment of the autoinhibitory domain (E2Pinter and E2Pactive) leads to a rigid-body movement of the P domain.
Extended Data Fig. 6 Processing pipeline for cryo-EM data of Drs2p–Cdc50p E2Pinhib.
a, Representative motion-corrected and dose-weighted micrograph (defocus 1.6 µm) of autoinhibited Drs2pΔN104–Cdc50p in LMNG, frozen at a concentration of 0.6 mg ml−1. b, Fourier power spectrum of the micrograph shown in a, as well as the fit from CTFFIND 4.1 through cisTEM, which extends to 3 Å. c, Colour code for the processing software. d, Data-processing workflow, indicating the number of particles that remained after each step at which particles were discarded. The densities resulting from 3D refinement are shown in grey, and relevant masks are light blue. The resolutions listed for 3D refinements are at FSC = 0.143.
Extended Data Fig. 7 Processing pipeline for cryo-EM data of Drs2p–Cdc50p E2Pinter.
a, Representative motion-corrected and dose-weighted micrograph (defocus 1.5 µm) of Drs2pΔN104–Cdc50p with an intact C terminus in LMNG, frozen at a concentration of 0.6 mg ml−1 in the presence of 75 µg ml−1 brain PI4P. b, Fourier power spectrum of micrograph in a, as well as the fit from CTFFIND 4.1 through cisTEM, which extends to 5 Å. c, Colour code for the processing software. d, Data-processing workflow, indicating the number of particles that remained after each step at which particles were discarded. The densities resulting from 3D refinement are shown in grey, and relevant masks are light blue. The resolutions listed for 3D refinements are at FSC = 0.143. The 3.4-Å refinement suggested a mixed state around TM10. To classify the structural heterogeneity that was caused by incomplete binding of PI4P, new ab initio references were generated in cryoSPARC, allowing for high similarity because the conformations were expected to be similar. Two different conformations resulted: the autoinhibited one and a PI4P-bound version. The autoinhibited conformation was identical to E2Pinhib and adding these particles to the E2Pinhib dataset did not improve the reconstruction. The PI4P-bound conformation was further refined in RELION.
Extended Data Fig. 8 Processing pipeline for cryo-EM data of Drs2p–Cdc50p E2Pactive.
a, Representative motion-corrected and dose weighted micrograph (defocus of 1.7 µm) of C-terminally truncated Drs2pΔNC–Cdc50p in LMNG, frozen at a concentration of 0.6 mg ml−1 in the presence of 75 µg ml−1 brain PI4P. b, Fourier power spectrum of the micrograph shown in a, as well as the fit from CTFFIND 4.1 through cisTEM, which extends to 3 Å. c, Colour code for the processing software. d, Data-processing workflow, indicating the number of particles that remained after each step at which particles were discarded. The densities resulting from 3D refinement are shown in grey, and relevant masks are light blue. The resolutions listed for 3D refinements are at FSC = 0.143.
Extended Data Fig. 9 Model validation and representative densities.
a, Cross-validation FSC curves for map-to-model fit produced by Mtriage60. Curves representing the model versus the full map are calculated based on the final model and the full, filtered and sharpened map that it was refined against. For the model versus half-maps, the model (before the final refinement) was refined against half-map 1 (filtered and sharpened as for the full map), and FSC curves were calculated using this refined model against each half-map. b, Representative densities from different areas of the three LocScale maps. Each segment is labelled with the residues that are shown, and demonstrates the quality of the map in specific areas. All densities are shown at a threshold of 1.5 r.m.s.d.
Supplementary information
Supplementary Discussion
Additional discussion on the similarity of P4-ATPases to known structures and the architecture of the complex with CDC50p. Supplementary Figure depicting sequence alignments.
Video 1
Cryo-EM density map. Final density map (with decreasing contour levels) and final model for E2Pinhib (Drs2p ΔN104/Cdc50p complex in the E2-BeF3- state and in the absence of PI4P). Domain colours same as in Fig. 1a.
Video 2
PI4P-mediated activation of Drs2p:Cdc50p. Release of autoinhibition triggers structural rearrangements in Drs2p leading to the opening of a cleft in the transmembrane domain suitable for the binding of the transport substrate. PI4P binding structures the C-terminus and destabilizes the autoinhibitory domain. Unstructured regions depicted by a dashed line. The movement of the autoinhibitory domain was manually modelled in Coot.
Rights and permissions
About this article
Cite this article
Timcenko, M., Lyons, J.A., Januliene, D. et al. Structure and autoregulation of a P4-ATPase lipid flippase. Nature 571, 366–370 (2019). https://doi.org/10.1038/s41586-019-1344-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1344-7
This article is cited by
-
Diverse roles of the metal binding domains and transport mechanism of copper transporting P-type ATPases
Nature Communications (2024)
-
TMEM30A is essential for hair cell polarity maintenance in postnatal mouse cochlea
Cellular & Molecular Biology Letters (2023)
-
P4-ATPase subunit Cdc50 plays a role in yeast budding and cell wall integrity in Candida glabrata
BMC Microbiology (2023)
-
Substrate binding-induced conformational transitions in the omega-3 fatty acid transporter MFSD2A
Nature Communications (2023)
-
Activation and substrate specificity of the human P4-ATPase ATP8B1
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.