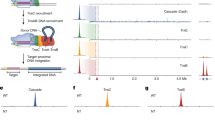
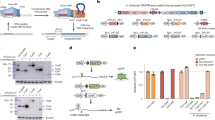
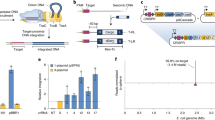
Abstract
Conventional CRISPR–Cas systems maintain genomic integrity by leveraging guide RNAs for the nuclease-dependent degradation of mobile genetic elements, including plasmids and viruses. Here we describe a notable inversion of this paradigm, in which bacterial Tn7-like transposons have co-opted nuclease-deficient CRISPR–Cas systems to catalyse RNA-guided integration of mobile genetic elements into the genome. Programmable transposition of Vibrio cholerae Tn6677 in Escherichia coli requires CRISPR- and transposon-associated molecular machineries, including a co-complex between the DNA-targeting complex Cascade and the transposition protein TniQ. Integration of donor DNA occurs in one of two possible orientations at a fixed distance downstream of target DNA sequences, and can accommodate variable length genetic payloads. Deep-sequencing experiments reveal highly specific, genome-wide DNA insertion across dozens of unique target sites. This discovery of a fully programmable, RNA-guided integrase lays the foundation for genomic manipulations that obviate the requirements for double-strand breaks and homology-directed repair.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Next-generation sequencing data are available in the National Center for Biotechnology Information Sequence Read Archive (BioProject Accession: PRJNA546035). Custom Python scripts used for the described data analyses are available online via GitHub (https://github.com/sternberglab/Klompe_etal_2019).
References
Thomas, C. M. & Nielsen, K. M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3, 711–721 (2005).
Soucy, S. M., Huang, J. & Gogarten, J. P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482 (2015).
Koonin, E. V. The turbulent network dynamics of microbial evolution and the statistical tree of life. J. Mol. Evol. 80, 244–250 (2015).
Toussaint, A. & Chandler, M. Prokaryote genome fluidity: toward a system approach of the mobilome. Methods Mol. Biol. 804, 57–80 (2012).
Dy, R. L., Richter, C., Salmond, G. P. C. & Fineran, P. C. Remarkable mechanisms in microbes to resist phage infections. Annu. Rev. Virol. 1, 307–331 (2014).
Hille, F. et al. The biology of CRISPR-Cas: backward and forward. Cell 172, 1239–1259 (2018).
Doron, S. et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 359, eaar4120 (2018).
Koonin, E. V., Makarova, K. S. & Wolf, Y. I. Evolutionary genomics of defense systems in archaea and bacteria. Annu. Rev. Microbiol. 71, 233–261 (2017).
Koonin, E. V. & Makarova, K. S. Mobile genetic elements and evolution of CRISPR-Cas systems: all the way there and back. Genome Biol. Evol. 9, 2812–2825 (2017).
Broecker, F. & Moelling, K. Evolution of immune systems from viruses and transposable elements. Front. Microbiol. 10, 51 (2019).
Kapitonov, V. V., Makarova, K. S. & Koonin, E. V. ISC, a novel group of bacterial and archaeal DNA transposons that encode Cas9 homologs. J. Bacteriol. 198, 797–807 (2016).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
Krupovic, M., Béguin, P. & Koonin, E. V. Casposons: mobile genetic elements that gave rise to the CRISPR-Cas adaptation machinery. Curr. Opin. Microbiol. 38, 36–43 (2017).
Peters, J. E., Makarova, K. S., Shmakov, S. & Koonin, E. V. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl Acad. Sci. USA 114, E7358–E7366 (2017).
Peters, J. E. Tn7. Microbiol. Spectr. 2, MDNA3-0010-2014 (2014).
Waddell, C. S. & Craig, N. L. Tn7 transposition: two transposition pathways directed by five Tn7-encoded genes. Genes Dev. 2, 137–149 (1988).
Lichtenstein, C. & Brenner, S. Unique insertion site of Tn7 in the E. coli chromosome. Nature 297, 601–603 (1982).
McKown, R. L., Orle, K. A., Chen, T. & Craig, N. L. Sequence requirements of Escherichia coli attTn7, a specific site of transposon Tn7 insertion. J. Bacteriol. 170, 352–358 (1988).
Parks, A. R. et al. Transposition into replicating DNA occurs through interaction with the processivity factor. Cell 138, 685–695 (2009).
McDonald, N. D., Regmi, A., Morreale, D. P., Borowski, J. D. & Boyd, E. F. CRISPR-Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC Genomics 20, 105 (2019).
Makarova, K. S., Wolf, Y. I. & Koonin, E. V. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J. 1, 325–336 (2018).
Rollins, M. F., Schuman, J. T., Paulus, K., Bukhari, H. S. T. & Wiedenheft, B. Mechanism of foreign DNA recognition by a CRISPR RNA-guided surveillance complex from Pseudomonas aeruginosa. Nucleic Acids Res. 43, 2216–2222 (2015).
Sarnovsky, R. J., May, E. W. & Craig, N. L. The Tn7 transposase is a heteromeric complex in which DNA breakage and joining activities are distributed between different gene products. EMBO J. 15, 6348–6361 (1996).
Stellwagen, A. E. & Craig, N. L. Gain-of-function mutations in TnsC, an ATP-dependent transposition protein that activates the bacterial transposon Tn7. Genetics 145, 573–585 (1997).
Haurwitz, R. E., Jinek, M., Wiedenheft, B., Zhou, K. & Doudna, J. A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329, 1355–1358 (2010).
May, E. W. & Craig, N. L. Switching from cut-and-paste to replicative Tn7 transposition. Science 272, 401–404 (1996).
Choi, K. Y., Spencer, J. M. & Craig, N. L. The Tn7 transposition regulator TnsC interacts with the transposase subunit TnsB and target selector TnsD. Proc. Natl Acad. Sci. USA 111, E2858–E2865 (2014).
Wiedenheft, B. et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl Acad. Sci. USA 108, 10092–10097 (2011).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012).
Wiedenheft, B. et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 477, 486–489 (2011).
Guo, T. W. et al. Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex. Cell 171, 414–426.e12 (2017).
Xue, C. & Sashital, D. G. Mechanisms of type I-E and I-F CRISPR-Cas systems in Enterobacteriaceae. EcoSal Plus 8, ESP-0008-2018 (2019).
Blosser, T. R. et al. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol. Cell 58, 60–70 (2015).
Cooper, L. A., Stringer, A. M. & Wade, J. T. Determining the specificity of cascade binding, interference, and primed adaptation in vivo in the Escherichia coli type I-E CRISPR-Cas system. MBio 9, e02100-17 (2018).
Rutkauskas, M. et al. Directional R-loop formation by the CRISPR-Cas surveillance complex cascade provides efficient off-target site rejection. Cell Reports 10, 1534–1543 (2015).
Luo, M. L. et al. The CRISPR RNA-guided surveillance complex in Escherichia coli accommodates extended RNA spacers. Nucleic Acids Res. 44, 7385–7394 (2016).
Goodman, A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009).
van Opijnen, T., Bodi, K. L. & Camilli, A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Methods 6, 767–772 (2009).
Wiles, T. J. et al. Combining quantitative genetic footprinting and trait enrichment analysis to identify fitness determinants of a bacterial pathogen. PLoS Genet. 9, e1003716 (2013).
Craig, N. L., Craigie, R., Gellert, M. & Lambowitz, A. M. Mobile DNA III (2014).
Stellwagen, A. E. & Craig, N. L. Avoiding self: two Tn7-encoded proteins mediate target immunity in Tn7 transposition. EMBO J. 16, 6823–6834 (1997).
Sobecky, P. A. & Hazen, T. H. Horizontal gene transfer and mobile genetic elements in marine systems. Methods Mol. Biol. 532, 435–453 (2009).
Makarova, K. S. Beyond the adaptive immunity: sub- and neofunctionalization of CRISPR–Cas systems and their components. Paper presented at: CRISPR 2018 Meeting; Jun 20; Vilnius, Lithuania. (2018).
Cheng, D. R., Yan, W. X. & Scott, D. A. Discovery of Type VI-D CRISPR-Cas Systems. Paper presented at: CRISPR 2018 Meeting; Jun 21; Vilnius, Lithuania. (2018).
Shmakov, S. et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 15, 169–182 (2017).
Dunbar, C. E. et al. Gene therapy comes of age. Science 359, eaan4672 (2018).
Gelvin, S. B. Integration of agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 51, 195–217 (2017).
Wurm, F. M. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 22, 1393–1398 (2004).
Kvaratskhelia, M., Sharma, A., Larue, R. C., Serrao, E. & Engelman, A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 42, 10209–10225 (2014).
Di Matteo, M., Belay, E., Chuah, M. K. & Vandendriessche, T. Recent developments in transposon-mediated gene therapy. Expert Opin. Biol. Ther. 12, 841–858 (2012).
Zelensky, A. N., Schimmel, J., Kool, H., Kanaar, R. & Tijsterman, M. Inactivation of Pol θ and C-NHEJ eliminates off-target integration of exogenous DNA. Nat. Commun. 8, 66 (2017).
Cox, D. B. T., Platt, R. J. & Zhang, F. Therapeutic genome editing: prospects and challenges. Nat. Med. 21, 121–131 (2015).
Pawelczak, K. S., Gavande, N. S., VanderVere-Carozza, P. S. & Turchi, J. J. Modulating DNA repair pathways to improve precision genome engineering. ACS Chem. Biol. 13, 389–396 (2018).
Schmidt, F., Cherepkova, M. Y. & Platt, R. J. Transcriptional recording by CRISPR spacer acquisition from RNA. Nature 562, 380–385 (2018).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448 (2018).
Yan, W. X. et al. Functionally diverse type V CRISPR-Cas systems. Science 363, 88–91 (2019).
Harrington, L. B. et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 362, 839–842 (2018).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Biswas, A., Gagnon, J. N., Brouns, S. J. J., Fineran, P. C. & Brown, C. M. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 10, 817–827 (2013).
Shevchenko, A., Tomas, H., Havlis, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protocols 1, 2856–2860 (2006).
Heidrich, N., Dugar, G., Vogel, J. & Sharma, C. M. Investigating CRISPR RNA biogenesis and function using RNA-seq. Methods Mol. Biol. 1311, 1–21 (2015).
Reiter, W. D., Palm, P. & Yeats, S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 17, 1907–1914 (1989).
Boyd, E. F., Almagro-Moreno, S. & Parent, M. A. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17, 47–53 (2009).
Acknowledgements
We thank M. I. Hogan for laboratory support, S. P. Chen and H. H. Wang for discussions, S. J. Resnick and A. Chavez for assistance with NGS experiments, R. Neme for assistance with NGS data analysis, L. F. Landweber for qPCR instrument access, the Department of Microbiology & Immunology for facilities and equipment support, the JP Sulzberger Columbia Genome Center for NGS support, and R. K. Soni and the Herbert Irving Comprehensive Cancer Center for proteomics support. Funding was provided by a generous start-up package from the Columbia University Irving Medical Center Dean’s Office and the Vagelos Precision Medicine Fund.
Author information
Authors and Affiliations
Contributions
S.E.K. and S.H.S. conceived of and designed the project. S.E.K. performed most transposition experiments, generated NGS libraries, and analysed the data. P.L.H.V. helped with cloning and transposition experiments, and performed computational analyses. T.S.H.-H. performed biochemical experiments. S.H.S., S.E.K. and all other authors discussed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
Columbia University has filed a patent application related to this work for which S.E.K. and S.H.S. are inventors. S.E.K. and S.H.S. are inventors on other patents and patent applications related to CRISPR–Cas systems and uses thereof. S.H.S. is a co-founder and scientific advisor to Dahlia Biosciences, and an equity holder in Dahlia Biosciences and Caribou Biosciences.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Transposition of the E. coli Tn7 transposon and genetic architecture of the Tn6677 transposon from V. cholerae.
a, Genomic organization of the native E. coli Tn7 transposon adjacent to its known attachment site (attTn7) within the glmS gene. b, Expression plasmid and donor plasmid for Tn7 transposition experiments. c, Genomic locus containing the conserved TnsD-binding site (attTn7), including the expected and alternative orientation Tn7 transposition products and PCR primer pairs to selectively amplify them. d, PCR analysis of Tn7 transposition, resolved by agarose gel electrophoresis. Amplification of rssA serves as a loading control; gel source data may be found in Supplementary Fig. 1. e, Sanger sequencing chromatograms of both upstream and downstream junctions of genomically integrated Tn7. f, Genomic organization of the native V. cholerae strain HE-45 Tn6677 transposon. Genes that are conserved between Tn6677 and the E. coli Tn7 transposon, and between Tn6677 and a canonical type I-F CRISPR–Cas system from P. aeruginosa28, are highlighted. The cas1 and cas2-3 genes, which mediate spacer acquisition and DNA degradation during the adaptation and interference stages of adaptive immunity, respectively, are missing from CRISPR–Cas systems encoded by Tn7-like transposons. Similarly, the tnsE gene, which facilitates non-sequence-specific transposition, is absent. The V. cholerae HE-45 genome contains another Tn7-like transposon (located within GenBank accession ALED01000025.1), which lacks an encoded CRISPR–Cas system and exhibits low sequence similarity to the Tn6677 transposon investigated in this study.
Extended Data Fig. 2 Analysis of E. coli cultures and strain isolates containing lacZ-integrated transposons.
a, Top, genomic locus targeted by crRNA-3 and crRNA-4, including both potential transposition products and the PCR primer pairs to selectively amplify them. Bottom, NGS analysis of the distance between the Cascade target site and transposon insertion site for crRNA-3 (left) and crRNA-4 (right), determined with two alternative primer pairs. b, Top, schematic of the lacZ locus with or without integrated transposon after transposition experiments with crRNA-4. T-LR and T-RL denote transposition products in which the transposon left end and right end are proximal to the target site, respectively. Primer pairs g and h (external–internal) selectively amplify the integrated locus, whereas primer pair i (external–external) amplifies both unintegrated and integrated loci. Bottom, PCR analysis of 10 colonies after 24-h growth on +IPTG plates (left) indicates that all colonies contain integration events in both orientations (primer pairs g and h), but with efficiencies sufficiently low that the unintegrated product predominates after amplification with primer pair i. After resuspending cells, allowing for an additional 18 h of clonal growth on −IPTG plates, and performing the same PCR analysis on 10 colonies (right), 3 out of 10 colonies now exhibit clonal integration in the T-LR orientation (compare primer pairs h and i). The remaining colonies show low-level integration in both orientations, which presumably occurred during the additional 18-h growth owing to leaky expression. These analyses indicate that colonies are genetically heterogeneous after growth on +IPTG plates, and that RNA-guided DNA integration only occurs in a proportion of cells within growing colonies. I, integrated product; U, unintegrated product. Asterisk denotes mispriming product also present in the negative (unintegrated) control. c, Photograph of LB-agar plate used for blue–white colony screening. Cells from IPTG-containing plates were replated on X-gal-containing plates, and white colonies expected to contain lacZ-inactivating transposon insertions were selected for further characterization. d, PCR analysis of E. coli strains identified by blue–white colony screening that contain clonally integrated transposons, as in b. e, Schematic of Sanger sequencing coverage across the lacZ locus for strains shown in d. f, PCR analysis of transposition experiment with crRNA-4 after serially diluting lysate from a clonally integrated strain with lysate from a control strain to simulate variable integration efficiencies, as in b. These experiments demonstrate that transposition products can be reliably detected by PCR with an external–internal primer pair at efficiencies above 0.5%, but that PCR bias leads to preferential amplification of the unintegrated product using the external-external primer pair at any efficiency substantially below 100%. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 3 Analysis of V. cholerae Cascade and TniQ–Cascade complexes.
a, Expression vectors for recombinant protein or ribonucleoprotein complex purification. b, Left, SDS–PAGE analysis of purified TniQ, Cascade and TniQ–Cascade complexes, highlighting protein bands excised for in-gel trypsin digestion and mass spectrometry analysis. Right, table listing E. coli and recombinant proteins identified from these data, and spectral counts of their associated peptides. Note that Cascade and TniQ–Cascade samples used for this analysis are distinct from the samples presented in Fig. 2. c, Size-exclusion chromatogram of the TniQ–Cascade co-complex on a Superose 6 10/300 column (left), and a calibration curve generated using protein standards (right). The measured retention time of TniQ–Cascade (maroon) is consistent with a complex having a molecular mass of approximately 440 kDa. d, RNase A and DNase I sensitivity of nucleic acids that co-purified with Cascade and TniQ–Cascade, resolved by denaturing urea–PAGE. e, TniQ, Cascade and a Cascade + TniQ binding reaction were resolved by size-exclusion chromatography (left), and indicated fractions were analysed by SDS–PAGE (right). Asterisk denotes an HtpG contaminant. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 Control experiments demonstrating efficient DNA targeting with Cas9 and P. aeruginosa Cascade.
a, Plasmid expression system for S. pyogenes (Spy) Cas9-sgRNA (type II-A, left) and P. aeruginosa Cascade (PaeCascade) and Cas2-3 (type I-F, right). The Cas2-3 expression plasmid was omitted from experiments described in Fig. 2e. b, Cell killing experiments using S. pyogenes Cas9-sgRNA (left) or PaeCascade and Cas2-3 (right), monitored by determining colony-forming units (CFU) after plasmid transformation. Complexes were programmed with guide RNAs that target the same genomic lacZ sites as with V. cholerae crRNA-3 and crRNA-4, such that efficient DNA targeting and degradation results in lethality and thus a drop in transformation efficiency. c, qPCR-based quantification of transposition efficiency from experiments using the V. cholerae transposon donor and TnsA-TnsB-TnsC, together with DNA targeting components comprising V. cholerae Cascade (Vch), P. aeruginosa Cascade (Pae) or S. pyogenes dCas9–RNA (dCas9). TniQ was expressed either on its own from pTnsABCQ or as a fusion to the targeting complex (pCas-Q) at the Cas6 C terminus (6), Cas8 N terminus (8), or dCas9 N or C terminus. The same sample lysates as in Fig. 2e were used. Data in b and c are shown as mean ± s.d. for n = 3 biologically independent samples.
Extended Data Fig. 5 qPCR-based quantification of RNA-guided DNA integration efficiencies.
a, Potential lacZ transposition products in either orientation for both crRNA-3 and crRNA-4, and qPCR primer pairs to selectively amplify them. b, Comparison of simulated integration efficiencies for T-LR and T-RL orientations, generated by mixing clonally integrated and unintegrated lysates in known ratios, versus experimentally determined integration efficiencies measured by qPCR. c, Comparison of simulated mixtures of bidirectional integration efficiencies for crRNA-4, generated by mixing clonally integrated and unintegrated lysates in known ratios, versus experimentally determined integration efficiencies measured by qPCR. d, RNA-guided DNA integration efficiency as a function of IPTG concentration for crRNA-3 and crRNA-4, measured by qPCR. Data in b and c are shown as mean ± s.d. for n = 3 biologically independent samples.
Extended Data Fig. 6 Influence of transposon end sequences on RNA-guided DNA integration.
a, Sequence (top) and schematic (bottom) of V. cholerae Tn6677 left- and right-end sequences. The putative TnsB-binding sites (blue) were determined based on sequence similarity to the TnsBbinding sites previously described14. The 8-bp terminal ends are shown in yellow, and the empirically determined minimum end sequences required for transposition are denoted by red dashed boxes. b, Integration efficiency with crRNA-4 as a function of transposon end length, as determined by qPCR. c, The relative fraction of both integration orientations as a function of transposon end length, determined by qPCR. ND, not determined. Data in b and c are shown as mean ± s.d. for n = 3 biologically independent samples.
Extended Data Fig. 7 Analysis of RNA-guided DNA integration for PAM-tiled crRNAs and extended spacer length crRNAs.
a, Integration site distribution for all crRNAs described in Fig. 3d, e having a normalized transposition efficiency more than 20%, determined by NGS. b, Integration site distribution for a crRNA containing mismatches at positions 29–32, compared with the distribution with crRNA-4, determined by NGS. c, The crRNA-4 spacer length was shortened or lengthened by 6-nucleotide increments, and the resulting integration efficiencies were determined by qPCR. Data are normalized to crRNA-4 and are shown as mean ± s.d. for n = 3 biologically independent samples. d, Integration site distribution for extended length crRNAs compared with the distribution with crRNA-4, determined by NGS.
Extended Data Fig. 8 Development and analysis of Tn-seq.
a, Schematic of the V. cholerae transposon end sequences. The 8-bp terminal sequence of the transposon is boxed and highlighted in light yellow. Mutations generated to introduce MmeI recognition sites are shown in red letters, and the resulting recognition site is highlighted in red. Cleavage by MmeI occurs 17–19 bp away from the transposon end, generating a 2-bp overhang. b, Comparison of integration efficiencies for the wild-type and MmeI-containing transposon donors, determined by qPCR. Labels on the x axis denote which plasmid was transformed last; we reproducibly observed higher integration efficiencies when pQCascade was transformed last (crRNA-4) than when pDonor was transformed last. The transposon containing an MmeI site in the transposon ‘right’ end (R∗-L pDonor) was used for all Tn-seq experiments. Data are mean ± s.d. for n = 3 biologically independent samples. c, Plasmid expression system for Himar1C9 and the mariner transposon. d, Scatter plot showing correlation between two biological replicates of Tn-seq experiments with the mariner transposon. Reads were binned by E. coli gene annotations, and a linear regression fit and Pearson linear correlation coefficient (r) are shown. e, Schematic of 100-bp binning approach used for Tn-seq analysis of transposition experiments with the V. cholerae transposon, in which bin 1 is defined as the first 100 bp immediately downstream (PAM-distal) of the Cascade target site. f, Scatter plots showing correlation between biological replicates of Tn-seq experiments with the V. cholerae transposon programmed with crRNA-4. All highly sampled reads fall within bin 1, but we also observed low-level but reproducible, long-range integration into 100-bp bins just upstream and downstream of the primary integration site (bins −1, 2 and 3). g, Scatter plot showing correlation between biological replicates of Tn-seq experiments with the V. cholerae transposon programmed with a non-targeting crRNA (crRNA-NT). h, Scatter plot showing correlation between biological replicates of Tn-seq experiments with the V. cholerae transposon expressing TnsA-TnsB-TnsC-TniQ but not Cascade. For f–h, bins are only plotted when they contain at least one read in either dataset.
Extended Data Fig. 9 Tn-seq data for additional crRNAs tested.
a, b, Genome-wide distribution of genome-mapping Tn-seq reads from transposition experiments with the V. cholerae transposon programmed with crRNAs 1–8 (a) and crRNAs 17–24 (b). The location of each target site is denoted by a maroon triangle. Dagger symbol indicates that the lacZ target site for crRNA-3 is duplicated within the λ DE3 prophage, as is the transposon integration site; Tn-seq reads for this dataset were mapped to both genomic loci for visualization purposes only, although we are unable to determine from which locus they derive. c, Analysis of integration site distributions for crRNAs 1–24 determined from the Tn-seq data; the distance between the Cascade target site and transposon insertion site is shown. Data for both integration orientations are superimposed, with filled blue bars representing the T-RL orientation and the dark outlines representing the T-LR orientation. Values in the top-right corner of each graph give the on-target specificity (%), calculated as the percentage of reads resulting from integration within 100 bp of the primary integration site, as compared with the total number of reads aligning to the genome; and the orientation bias (X:Y), calculated as the ratio of reads for the T-RL orientation to reads for the T-LR orientation. Most crRNAs favour integration in the T-RL orientation 49–50 bp downstream of the Cascade target site. crRNA-21 is greyed out because the expected primary integration site is present in a repetitive stretch of DNA that does not allow us to map the reads confidently. Asterisks denote samples for which more than 1% of the genome-mapping reads could not be uniquely mapped.
Extended Data Fig. 10 Bacterial transposons also contain type V-U5 CRISPR–Cas systems encoding C2c5.
Representative genomic loci from various bacterial species containing identifiable transposon left and right ends (blue boxes, L and R), genes with homology to tnsB-tnsC-tniQ (shades of yellow), CRISPR arrays (maroon), and the CRISPR-associated gene c2c5 (blue). The example from Hassallia byssoidea (top) highlights the target-site duplication and terminal repeats, as well as genes found within the cargo portion of the transposon. As with the type I CRISPR–Cas system-containing Tn7-like transposons, type V CRISPR–Cas system-containing transposons appear to preferentially contain genes associated with innate immune system functions, such as restriction-modification systems. c2c5 genes are frequently flanked by the predicted transcriptional regulator, merR (light blue), and the C2c5-containing transposons appear to usually fall just upstream of tRNA genes (green), a phenomenon that has also been observed for other prokaryotic integrative elements62,63. Analysis of 50 spacers from the 8 CRISPR arrays shown with CRISPRTarget59 revealed 6 spacers with imperfectly matching targets (average of 6 mismatches), none of which mapped to bacteriophages, plasmids, or to the same bacterial genome containing the transposon itself. Whether C2c5 also mediates RNA-guided DNA integration awaits future experimentation.
Supplementary information
Supplementary Note
Nomenclature for transposons and CRISPR-Cas systems described in this study.
Supplementary Figures
This file contains Supplementary Figures 1-8 including legends.
Supplementary Table 1
Description and sequence of plasmids used in this study.
Supplementary Table 2
Gene and protein sequences for the Vibrio cholerae RNA-guided DNA integration machinery used in this study. ∗ The V. cholerae HE-45 genome contains another Tn7-like transposon (GenBank accession ALED01000025.1), which lacks an encoded CRISPR–Cas system and exhibits low sequence similarity to the transposon investigated in this study. † The gene sequences shown are copied from the Vibrio cholerae HE-45 genome. Actual sequences used in this study contained additional silent point mutations for cloning purposes, and can be found in Supplementary Table 1. ‡ The protein sequences shown are full-length translations from the Vibrio cholerae HE-45 genome. TnsA in our experiments contained an additional alanine residue after the N-terminal methionine. § Cas8 is a Cas8-Cas5 fusion protein, as described in the main text.
Supplementary Table 3
Guide RNAs and genomic target sites used in this study. ∗ Coordinates are for the E. coli BL21(DE3) genome (GenBank accession CP001509). † PAM sequences denote the 2 nucleotides immediately 5’ of the target (V. cholerae and P. aeruginosa Cascade) or 3 nucleotides immediately 3’ of the target (S. pyogenes Cas9) on the non-target strand.
Supplementary Table 4
Next-generation sequencing library statistics.
Supplementary Table 5
Oligonucleotides used for PCR, qPCR, and NGS experiments in this study.
Rights and permissions
About this article
Cite this article
Klompe, S.E., Vo, P.L.H., Halpin-Healy, T.S. et al. Transposon-encoded CRISPR–Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019). https://doi.org/10.1038/s41586-019-1323-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1323-z
This article is cited by
-
Genomic language model predicts protein co-regulation and function
Nature Communications (2024)
-
CRISPR technologies for genome, epigenome and transcriptome editing
Nature Reviews Molecular Cell Biology (2024)
-
Precise genome-editing in human diseases: mechanisms, strategies and applications
Signal Transduction and Targeted Therapy (2024)
-
Targeted DNA integration in human cells without double-strand breaks using CRISPR-associated transposases
Nature Biotechnology (2024)
-
Bacterial genome engineering using CRISPR-associated transposases
Nature Protocols (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.