Abstract
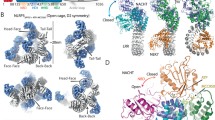
The NLRP3 inflammasome can be activated by stimuli that include nigericin, uric acid crystals, amyloid-β fibrils and extracellular ATP. The mitotic kinase NEK7 licenses the assembly and activation of the NLRP3 inflammasome in interphase. Here we report a cryo-electron microscopy structure of inactive human NLRP3 in complex with NEK7, at a resolution of 3.8 Å. The earring-shaped NLRP3 consists of curved leucine-rich-repeat and globular NACHT domains, and the C-terminal lobe of NEK7 nestles against both NLRP3 domains. Structural recognition between NLRP3 and NEK7 is confirmed by mutagenesis both in vitro and in cells. Modelling of an active NLRP3–NEK7 conformation based on the NLRC4 inflammasome predicts an additional contact between an NLRP3-bound NEK7 and a neighbouring NLRP3. Mutations to this interface abolish the ability of NEK7 or NLRP3 to rescue NLRP3 activation in NEK7-knockout or NLRP3-knockout cells. These data suggest that NEK7 bridges adjacent NLRP3 subunits with bipartite interactions to mediate the activation of the NLRP3 inflammasome.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
References
Lamkanfi, M. & Dixit, V. M. Mechanisms and functions of inflammasomes. Cell 157, 1013–1022 (2014).
Broz, P. & Dixit, V. M. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16, 407–420 (2016).
Guo, H., Callaway, J. B. & Ting, J. P. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21, 677–687 (2015).
Shi, J., Gao, W. & Shao, F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 42, 245–254 (2017).
Liu, X. et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016).
Hu, Z. et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science 341, 172–175 (2013).
Lu, A. et al. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 156, 1193–1206 (2014).
Lu, A. et al. Molecular basis of caspase-1 polymerization and its inhibition by a new capping mechanism. Nat. Struct. Mol. Biol. 23, 416–425 (2016).
Lu, A. et al. Plasticity in PYD assembly revealed by cryo-EM structure of the PYD filament of AIM2. Cell Discov. 1, 15013 (2015).
Rathinam, V. A., Vanaja, S. K. & Fitzgerald, K. A. Regulation of inflammasome signaling. Nat. Immunol. 13, 333–342 (2012).
Muñoz-Planillo, R. et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 (2013).
Chen, J. & Chen, Z. J. PtdIns4P on dispersed trans-Golgi network mediates NLRP3 inflammasome activation. Nature 564, 71–76 (2018).
He, Y., Zeng, M. Y., Yang, D., Motro, B. & Núñez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 530, 354–357 (2016).
Shi, H. et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17, 250–258 (2016).
Schmid-Burgk, J. L. et al. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291, 103–109 (2016).
Shi, H., Murray, A. & Beutler, B. Reconstruction of the mouse inflammasome system in HEK293T cells. Bio Protoc. 6, e1986 (2016).
Coll, R. C. et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 (2015).
Dar, A. C., Dever, T. E. & Sicheri, F. Higher-order substrate recognition of eIF2α by the RNA-dependent protein kinase PKR. Cell 122, 887–900 (2005).
Hu, Z. et al. Structural and biochemical basis for induced self-propagation of NLRC4. Science 350, 399–404 (2015).
Zhang, L. et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science 350, 404–409 (2015).
Maekawa, S., Ohto, U., Shibata, T., Miyake, K. & Shimizu, T. Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 7, 11813 (2016).
Yang, X. et al. Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res. 28, 35–47 (2018).
Tenthorey, J. L. et al. The structural basis of flagellin detection by NAIP5: a strategy to limit pathogen immune evasion. Science 358, 888–893 (2017).
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 (2015).
Richards, M. W. et al. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol. Cell 36, 560–570 (2009).
Qu, Y. et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 (2012).
Suzuki, S. et al. Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcδ. PLoS Pathog. 10, e1003926 (2014).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Haq, T. et al. Mechanistic basis of Nek7 activation through Nek9 binding and induced dimerization. Nat. Commun. 6, 8771 (2015).
Spalinger, M. R. et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Invest. 126, 1783–1800 (2016).
Hafner-Bratković, I. et al. NLRP3 lacking the leucine-rich repeat domain can be fully activated via the canonical inflammasome pathway. Nat. Commun. 9, 5182 (2018).
Moghaddas, F. et al. Autoinflammatory mutation in NLRC4 reveals a leucine-rich repeat (LRR)-LRR oligomerization interface. J. Allergy Clin. Immunol. 142, 1956–1967.e6 (2018).
Touitou, I. et al. Infevers: an evolving mutation database for auto-inflammatory syndromes. Hum. Mutat. 24, 194–198 (2004).
Duncan, J. A. et al. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl Acad. Sci. USA 104, 8041–8046 (2007).
Jiang, X. & Wang, X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to Apaf-1. J. Biol. Chem. 275, 31199–31203 (2000).
Song, N. & Li, T. Regulation of NLRP3 inflammasome by phosphorylation. Front. Immunol. 9, 2305 (2018).
Barry, R. et al. SUMO-mediated regulation of NLRP3 modulates inflammasome activity. Nat. Commun. 9, 3001 (2018).
Py, B. F., Kim, M. S., Vakifahmetoglu-Norberg, H. & Yuan, J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol. Cell 49, 331–338 (2013).
Mangan, M. S. J. et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 17, 688 (2018).
Khan, M., Subramaniam, R. & Desveaux, D. Of guards, decoys, baits and traps: pathogen perception in plants by type III effector sensors. Curr. Opin. Microbiol. 29, 49–55 (2016).
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Wu, J. et al. Massively parallel unsupervised single-particle cryo-EM data clustering via statistical manifold learning. PLoS ONE 12, e0182130 (2017).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Delano, W. L. The PyMol Molecular Graphics System https://pymol.org/2/ (2002).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W320 (2014).
Acknowledgements
We thank the University of Massachusetts Cryo-EM Core Facility and the National Cryo-EM Facility at the National Cancer Institute for 200-keV data collection, the Electron Microscopy Laboratory and Cryo-EM Platform at Peking University for 300-keV data collection, and C. Xu, K. Song, K. Lee, X. Li, Y. Ma and D. Yu for technical support. This work was funded in part by the US NIH grant DP1HD087988 (H.W.), R01Al124491 (H.W.), R01AI06331 (G.N.), an Intel Corporation academic grant (Y.M.), the Thousand Talents Plan of China (Y.M.), National Natural Science Foundation of China grant 11774012 (Y.M.) and Natural Science Foundation of Beijing City grant Z180016 (Y.M.). Data processing was performed in part in the Sullivan cluster, which is funded in part by a gift from Mr. and Mrs. D. J. Sullivan Jr, and in the Weiming No.1 and Life Science No. 1 High-Performance Computing Platform at Peking University. We thank H. Leung (PCMM, BCH confocal core facility manager).
Author information
Authors and Affiliations
Contributions
H.W., L.W. and H.S. conceived the study. L.W. designed constructs. L.W., Q.Q. and H.S. purified the complexes, and H.S., L.W. and Q.Q. made cryo-grids for data collection. H.S., L.W., W.L.W., Q.Q., Z.W. and Y.M. collected data. H.S., W.L.W. and Q.Q. analysed cryo-EM data, and H.W. and Y.M. supervised data processing. H.S. performed initial model building and refinement, and W.L.W. and Y.M. performed additional model fitting and refinement. H.S. designed mutants for in vitro and cell-based assays. V.G.M. performed all cell-based assays. L.A. and A.V.H. performed NEK7-mutant assays. G.N. provided reagents for cell-based assays and valuable discussions. H.W., H.S. and Y.M. wrote the manuscript, and all authors provided comments on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
L.W. and A.V.H. are employees, and H.W. is co-founder, of SMOC Therapeutics; the other authors declare no competing interests.
Additional information
Peer Reviewer information:
Nature thanks Eicke Latz, Edward Miao and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 NLRP3–NEK7 protein purification and characterization.
a, Raw traces (of three independent replicates) of microscale thermophoresis measurements corresponding to titration of NEK7 against Alexa Fluor 488-labelled NLRP3. b–d, Representative thermal denaturation curves of NLRP3 (b), NEK7 (c) and complex (d), alone and in the presence of ADP and/or MCC950. The peak minima of the derivative curves correspond to the protein melting temperatures (Tm) (repeated ≥ 3 times). e, Gel filtration profile of wild-type NEK7 (monomer) and engineered NEK7 dimer on Superdex 200 column (repeated ≥ 5 times). f, Molecular masses of NLRP3–NEK7 dimer and monomer complexes, measured by in-line MALS.
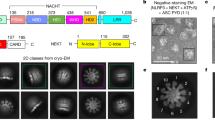
Extended Data Fig. 2 Analysis and workflow for cryo-EM data collected on a Talos Arctica and on a Titan Krios.
a, A negative-staining electron microscopy image (top, one among a few hundred images) and a cryo-EM micrograph (bottom, one among a few thousand images) of the NLRP3–NEK7 complex from a Talos Arctica. Scale bars, 50 nm. b, Workflow of cryo-EM data analysis of the Arctica data, performed in RELION 3.053 and cisTEM46. c, Gold-standard FSC curve between two half maps from the Arctica data. d, Local resolution estimation of the Arctica map generated by ResMap41, coloured on the cryo-EM density (8σ). The highest resolution is observed where NEK7 interacts with NLRP3. e, A cryo-EM micrograph of the NLRP3–NEK7 complex from a Titan Krios (one among more than 10,000 images). Scale bar, 50 nm. The inset shows the modelled (left) and actual (right) Thon rings. f, Workflow of cryo-EM data analysis of the Krios data, done in RELION 3.053 and ROME 1.148. The top right insets show the orientation distributions of the particles from the dataset of tilt 0° and tilt 20°. g, Gold-standard FSC curves between two half maps from the Krios data with mask (orange) and without mask (green), and between map and model (blue).
Extended Data Fig. 3 Local density fitted with NLRP3 and NEK7.
All magnified views are labelled with domain names and selected segment residue numbers. Densities are shown at 3σ.
Extended Data Fig. 4 Predicted and cryo-EM-derived secondary structures of the NLRP3–LRR domains aligned to NLRP3 and NAIP5 sequences.
Twelve LRRs in NLRP3 were predicted by the Phyre2 server24.
Extended Data Fig. 5 The structure of the NLRP3–NEK7 complex, showing the modelled full-length NEK7 and NLRP3 domain comparisons.
a, Two views of the NLRP3–NEK7 complex structure with the NEK7 N-lobe (grey) modelled from the NEK7 crystal structure (PDB code 2WQM). b, The disordered activation loop, including S195 in the NLRP3–NEK7 complex, which is not involved in NLRP3 interaction. c, NBD, HD1, WHD and HD2 of NLRP3, NOD2 (PDB code 5IRL) and NLRC4 (PDB code 4KXF) superimposed and shown side-by-side for comparison. d, e, NOD2 (d) and NLRC4 (e) structures6,21 in inactive conformations and in the orientation of NLRP3 (shown in a) by superposing on the NBD and HD1 domains. The location of phosphorylated S533 of NLRC4 is labelled.
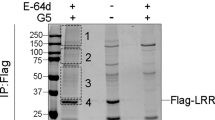
Extended Data Fig. 8 Structural analysis, mutagenesis tests and overlap between the NEK9- and NLRP3-binding sites on NEK7.
a, Position of the bound SO4 ion as in the NEK7 crystal structure25, superimposed here in the structure of the NLRP3–NEK7 complex. b, Cryo-EM densities (5σ) at the NLRP3–NEK7 interfaces. Views correspond to those of Fig. 3b, c. c, Amylose pulldown of wild-type and mutant NEK7 without NLRP3, showing input and precipitate gels, which serve as negative controls for Fig. 3d. Experiments were repeated 3–6 times. d, Input gel for amylose pulldown of wild-type and mutant NEK7 shown in Fig. 3d. Experiments were repeated 3–6 times. e, Alternative calculation of the percentage of NEK7 pulldown relative to wild type, by subtracting the NEK7/NLRP3 ratio in the absence of NEK7 from the observed NEK7/NLRP3 ratio (shown as mean ± s.d. for n = 3–6 experiments). f, Cytoplasmic LDH released into the supernatant, quantified by comparison with total intracellular LDH of untreated cells (triton-X-lysed). Data are presented as mean ± s.d. for n = 3 replicates from 2 independent experiments. g, Top, mapping the NEK9-binding site onto the structure of the NEK7–NEK9 complex (PDB code 5DE2)29. NEK7 and NEK9 are shown in yellow and cyan, respectively, with the NEK9-binding residues of NEK7 highlighted in red. Bottom, a rainbow-coloured NEK7 structure showing that the NEK9-binding residues are from the first part of the NEK7 C-lobe. h, Mapping the NLRP3-binding site (red) onto NEK7 in the NLRP3–NEK7 structure. The NLRP3-binding site overlaps with the NEK9-binding site on NEK7. i, Superposition of NEK7 back-to-back dimer (yellow and pink) in the NEK7–NEK9 complex structure (PDB code 5DE2)29 onto the NEK7 monomer in the NLRP3–NEK7 structure. The pink monomer in the NEK7 dimer clashes with NLRP3, which suggests that NEK7 dimerization cannot occur in the NLRP3–NEK7 complex. j, NLRP3-knockout iBMDMs were reconstituted with wild-type or mutant human mNG-tagged NLRP3, primed by LPS (4 h) and stimulated by nigericin (30 min). LDH release was analysed as in f. Dots, individual data points. Data are presented as mean ± s.d. for n = 3 replicates from 2 independent experiments. k, LRR phosphorylation at Y85930, which might cause steric and charge repulsion with NEK7. l, NLRP3 activation model and NEK7 interactions. The hypothetical ~90° rotation of the NBD and HD1 of NLRP3 upon activation moves the NBD away from the NEK7 interaction region (indicated by a box).
Extended Data Fig. 9 NLRP3–NEK7 interaction in trans and NLRP3 CAPS mutations.
a, Cryo-EM density (5σ) fitted to the oligomeric model, zoomed in at an NLRP3–NEK7 interface in trans. b, Immunofluorescence imaging for assessing ASC speck formation. NLRP3-knockout and NEK7-knockout iBMDMs were reconstituted with wild-type and mutant constructs as depicted. ASC speck formation upon nigericin activation was analysed by anti-ASC antibody. Nucleus was stained by Hoechst 33342. Scale bar, 10 μm. Images are representative of two independent experiments. c, An active NLRC4 oligomer structure20 shown for the LRR–LRR interaction. d, Magnified view showing the detailed LRR–LRR interaction in trans boxed in c. e, Mapping of pathogenic disease mutations onto the NLRP3 structure. CAPS-associated pathogenic mutations derived from the Infevers database33 are shown with their predicted effect, based on the NLRP3 structure. Green, disruption of inter-domain interactions in the inactive conformation; mauve, change of local conformation by mutating residues buried within the domains; yellow, alteration in key residues in the Walker B motif; grey, enhancement of NEK7 binding. CINCA, chronic infantile neurological cutaneous articular; NOMID, neonatal-onset multisystem inflammatory disorder; FCAS, familial cold auto-inflammatory syndrome.
Supplementary information
Supplementary Figure
This file contains Source Data Gels.
Video 1
Video depicts the overall structure of NLRP3-NEK7 bound with ADP. Domains are colour coded as described in the main text.
Video 2
Video depicts the NLRC4 modelled activation movement of NBD and HD1 domains of NRLP3 to bring it in an active conformation. The domains are colours as described in the main text
Rights and permissions
About this article
Cite this article
Sharif, H., Wang, L., Wang, W.L. et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 570, 338–343 (2019). https://doi.org/10.1038/s41586-019-1295-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1295-z
This article is cited by
-
Structural basis for the oligomerization-facilitated NLRP3 activation
Nature Communications (2024)
-
Structural basis of the subcortical maternal complex and its implications in reproductive disorders
Nature Structural & Molecular Biology (2024)
-
Mechanistic insights from inflammasome structures
Nature Reviews Immunology (2024)
-
Drugging the NLRP3 inflammasome: from signalling mechanisms to therapeutic targets
Nature Reviews Drug Discovery (2024)
-
Aiouea padiformis extract exhibits anti-inflammatory effects by inhibiting the ATPase activity of NLRP3
Scientific Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.