Abstract
Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci in prokaryotes are composed of 30–40-base-pair repeats separated by equally short sequences of plasmid and bacteriophage origin known as spacers1,2,3. These loci are transcribed and processed into short CRISPR RNAs (crRNAs) that are used as guides by CRISPR-associated (Cas) nucleases to recognize and destroy complementary sequences (known as protospacers) in foreign nucleic acids4,5. In contrast to most Cas nucleases, which destroy invader DNA4,5,6,7, the type VI effector nuclease Cas13 uses RNA guides to locate complementary transcripts and catalyse both sequence-specific cis- and non-specific trans-RNA cleavage8. Although it has been hypothesized that Cas13 naturally defends against RNA phages8, type VI spacer sequences have exclusively been found to match the genomes of double-stranded DNA phages9,10, suggesting that Cas13 can provide immunity against these invaders. However, whether and how Cas13 uses its cis- and/or trans-RNA cleavage activities to defend against double-stranded DNA phages is not understood. Here we show that trans-cleavage of transcripts halts the growth of the host cell and is sufficient to abort the infectious cycle. This depletes the phage population and provides herd immunity to uninfected bacteria. Phages that harbour target mutations, which easily evade DNA-targeting CRISPR systems11,12,13, are also neutralized when Cas13 is activated by wild-type phages. Thus, by acting on the host rather than directly targeting the virus, type VI CRISPR systems not only provide robust defence against DNA phages but also prevent outbreaks of CRISPR-resistant phage.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The L. seeligeri RR4 and L. ivanovii RR3 genome sequences, along with raw reads from the spacer library deep sequencing, paired-end RNA-seq, and 5′ end mapping have been deposited in the Sequence Read Archive under BioProject accession number PRJNA512236. Lists of strains, plasmids, and oligonucleotides used in this study are available in Supplementary Information 5.
Code availability
Custom scripts used in analysis of spacer library data as well as RNA 5′ end mapping data are available upon request.
References
Mojica, F. J., Díez-Villaseñor, C., García-Martínez, J. & Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 60, 174–182 (2005).
Bolotin, A., Quinquis, B., Sorokin, A. & Ehrlich, S. D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151, 2551–2561 (2005).
Pourcel, C., Salvignol, G. & Vergnaud, G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151, 653–663 (2005).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Brouns, S. J. et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321, 960–964 (2008).
Marraffini, L. A. & Sontheimer, E. J. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322, 1843–1845 (2008).
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Smargon, A. A. et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell 65, 618–630.e617 (2017).
Yan, W. X. et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell 70, 327–339.e325 (2018).
Deveau, H. et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 190, 1390–1400 (2008).
van Houte, S. et al. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388 (2016).
Pyenson, N. C., Gayvert, K., Varble, A., Elemento, O. & Marraffini, L. A. Broad targeting specificity during bacterial type III CRISPR–Cas immunity constrains viral escape. Cell Host Microbe 22, 343–353.e343 (2017).
Meeske, A. J. & Marraffini, L. A. RNA guide complementarity prevents self-targeting in type VI CRISPR systems. Mol. Cell 71, 791–801.e793 (2018).
Westra, E. R. et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol. Cell 46, 595–605 (2012).
Koonin, E. V. & Zhang, F. Coupling immunity and programmed cell suicide in prokaryotes: life-or-death choices. BioEssays 39, 1–9 (2017).
Parreira, R., Ehrlich, S. D. & Chopin, M. C. Dramatic decay of phage transcripts in lactococcal cells carrying the abortive infection determinant AbiB. Mol. Microbiol. 19, 221–230 (1996).
Fineran, P. C. et al. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl Acad. Sci. USA 106, 894–899 (2009).
Short, F. L. et al. Selectivity and self-assembly in the control of a bacterial toxin by an antitoxic noncoding RNA pseudoknot. Proc. Natl Acad. Sci. USA 110, E241–E249 (2013).
Shiloh, M. U., Ruan, J. & Nathan, C. Evaluation of bacterial survival and phagocyte function with a fluorescence-based microplate assay. Infect. Immun. 65, 3193–3198 (1997).
Watson, B. N. J., Staals, R. H. J. & Fineran, P. C. CRISPR–Cas-mediated phage resistance enhances horizontal gene transfer by transduction. MBio 9, e02406-17 (2018).
Payne, P., Geyrhofer, L., Barton, N. H. & Bollback, J. P. CRISPR-based herd immunity can limit phage epidemics in bacterial populations. eLife 7, e32035 (2018).
Klumpp, J. et al. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190, 5753–5765 (2008).
Rostøl, J. T. & Marraffini, L. (Ph)ighting phages: how bacteria resist their parasites. Cell Host Microbe 25, 184–194 (2019).
Yan, W. X. et al. Functionally diverse type V CRISPR–Cas systems. Science 363, 88–91 (2019).
Zetsche, B. et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR–Cas system. Cell 163, 759–771 (2015).
Hynes, A. P., Villion, M. & Moineau, S. Adaptation in bacterial CRISPR–Cas immunity can be driven by defective phages. Nat. Commun. 5, 4399 (2014).
Lauer, P., Chow, M. Y., Loessner, M. J., Portnoy, D. A. & Calendar, R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184, 4177–4186 (2002).
Rocourt, J., Schrettenbrunner, A., Hof, H. & Espaze, E. P. [New species of the genus Listeria: Listeria seeligeri]. Pathol. Biol. (Paris) 35, 1075–1080 (1987).
Lemaître, J. P., Duroux, A., Pimpie, R., Duez, J. M. & Milat, M. L. Listeria phage and phage tail induction triggered by components of bacterial growth media (phosphate, LiCl, nalidixic acid, and acriflavine). Appl. Environ. Microbiol. 81, 2117–2124 (2015).
Loessner, M. J., Inman, R. B., Lauer, P. & Calendar, R. Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes: implications for phage evolution. Mol. Microbiol. 35, 324–340 (2000).
Acknowledgements
We thank all members of the Marraffini laboratory for advice and encouragement, A. Varble for discussions, and J. T. Rostøl for critical reading of the manuscript. L. seeligeri RR4 and L. ivanovii RR3 were gifts from J.-P. Lemaître. Support for this work comes from the National Institute of Health Director’s Pioneer Award 1DP1GM128184-01 (to L.A.M.). L.A.M. is an investigator of the Howard Hughes Medical Institute. A.J.M. is a Helen Hay Whitney postdoctoral fellow.
Peer review information
Nature thanks Peter Fineran, Edze Westra and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Experiments were designed by A.J.M. and L.A.M. A.J.M. conducted spacer library construction and testing, all RNA sequencing, cell dormancy experiments, phage mutant construction, and escaper cross-protection assays, as well analysis of all next-generation sequencing data. S.N.-H. assisted with sequencing the L. seeligeri RR4 and L. ivanovii RR3 genomes and initial testing of spacers in ϕRR4 immunity. The paper was written by A.J.M. and L.A.M.
Corresponding authors
Ethics declarations
Competing interests
L.A.M. is a cofounder and Scientific Advisory Board member of Intellia Therapeutics, and a co-founder of Eligo Biosciences. The other authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
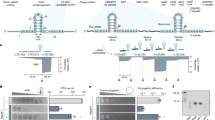
Extended Data Fig. 1 Listeria phage infection model for studying type VI-A CRISPR immunity.
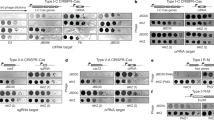
a, Diagram of the ϕRR4 genome, with individual genes depicted within the anti-CRISPR, early lytic, late lytic, and lysogenic regions. L. seeligeri ATCC35967 harbours a five-spacer type VI-A CRISPR locus29, but phages that infect this strain have not yet been identified. We sequenced the genome of L. seeligeri RR430 and found that it contains a 42-kb prophage, ϕRR4, that is similar to the A118 listeriophage31. Although ϕRR4 particles induced from the lysogen did not infect L. seeligeri ATCC35967, ϕRR4 propagated in the closely related L. ivanovii RR3 strain (99.2% 16S rRNA identity)30. b, The type VI-A CRISPR locus of L. seeligeri ATCC35967 was inserted into the tRNAArg gene of L. ivanovii RR3 using the vector pAM125, generating L. ivanovii ΩCRISPRVI. Different strains with either the five spacers naturally present in this system (spc1–5), individual spacers matching the genome of ϕRR4 (spcA, spcE, spcL), or a ϕRR4 spacer library (spc lib), were generated. For the latter, 41,276 ϕRR4-matching spacers were selected, tiled every two nucleotides across the phage genome, with both strands equally represented. c, Test of type VI-A anti-plasmid immunity in L. ivanovii ΩCRISPRVI(spc1–5). Plasmids with spc2 or spc4 targets in the chloramphenicol resistance cassette were conjugated into L. ivanovii RR3 or ΩCRISPRVI(spc1–5) and transconjugants selected on nalidixic acid and chloramphenicol. Transconjugants that received an empty vector lacking a target sequence are shown as a negative control (none). Representative of two biological replicates. d, Prevention of ϕRR4 lytic infection by the type VI spacer library. Spacer library cells (yellow–orange gradient) or cells lacking CRISPR (grey) were infected with ϕRR4 at OD600 = 0.1, MOI = 1, and OD600 was monitored over time. Representative of two biological replicates. e, One-hundred-base-pair sliding window average spacer enrichment ratio (post-infection/pre-infection spacer abundance) for spacers targeting the top (orange) and bottom (brown) strands.
Extended Data Fig. 2 ϕRR4 transcriptome and enrichment of corresponding targeting spacers.
a, RNA-seq over the course of ϕRR4 infection. Wild-type L. ivanovii RR3 was infected with ϕRR4 at an MOI of 1 and samples were collected for transcriptomic analysis by paired-end RNA-seq at the indicated time points. Reads were mapped to the ϕRR4 genome and normalized to the total reads per sample. Orange, top strand; brown, bottom strand. Representative of two biological replicates. b, Spacer enrichment correlates with target transcription, with no additional protection conferred above a critical expression threshold. Spacer abundance in the library was assessed before infection as well as 5 h after infection with ϕRR4 at an MOI of 1. Spacer enrichment distributions are shown, with individual histograms representing different tiers of target transcript abundance for the corresponding protospacer.
Extended Data Fig. 3 Cas13a-mediated cleavage of phage and host transcripts detected by RNA-seq.
a, Abundance of phage transcripts assessed by conventional paired-end RNA-seq 1.75 h after infection with ϕRR4 in L. ivanovii RR3 wild-type, ΩCRISPRVI(spcE) or ΩCRISPRVI(spcL) strains. Reads were mapped to the ϕRR4 genome and normalized to the abundance of a spike-in RNA. Both ΩCRISPRVI(spcE) and ΩCRISPRVI(spcL) targeting result in elevated early transcript cleavage products (Fig. 2a, b) and a reduction in late transcript abundance. Orange, top strand; brown, bottom strand. Representative of two biological replicates. b, L. ivanovii host mRNA cleavage detected by 5′ end mapping in L. ivanovii RR3 wild-type (grey) and ΩCRISPRVI(spcE) (green) strains 1.75 h after infection with ϕRR4. The height of each peak represents the detected abundance of the corresponding mRNA 5′ end. Grey arrowheads, TSSs. Four regions of the genome are depicted: murA1, ftsEX/iap, isdCD, division and cell wall (dcw) cluster. Abundant intragenic cleavage products are generated in the ΩCRISPRVI(spcE) strain. Representative of two biological replicates. c, The four genomic regions in b shown for the native type VI CRISPR host L. seeligeri, wild-type (red) and ∆CRISPR (grey), 15 min after aTc-mediated induction of a target transcript. The dcw cluster is broken into two operons in L. seeligeri. Representative of two biological replicates.
Extended Data Fig. 4 Trans-RNase activity is sufficient to limit growth of both ϕRR4 phage and ΩCRISPRVI host.
a, L. ivanovii RR3 and ΩCRISPRVI(spcA, spcE or spcL) strains at OD600 = 0.05 were infected with ϕRR4 at an MOI of 1 and growth was monitored over 24 h. Each curve represents the mean ± s.e.m. of three biological replicates. b, Quantification of ϕRR4 infective centres over time on wild-type L. ivanovii RR3. Cells were infected with ϕRR4 at an MOI of 0.1 and allowed to adsorb for 5 min, and then cells were washed three times to remove free phage. Infective centres were counted every 30 min by counting plaque-forming units on a lawn of phage-susceptible RR3 cells. Each data point represents the mean ± s.e.m. of three biological replicates. c, Survival of the indicated strains during ϕRR4 infection at an MOI of 2. CFU titres were measured before infection (P) and 4 h after infection (IN) or mock infection (UN). Each bar represents the mean ± s.e.m. of three biological replicates.
Extended Data Fig. 5 Activation of Cas13a induces reversible dormancy of host cells.
a, Growth arrest (measured as culture OD600) induced by target transcription in wild-type L. seeligeri (but not the ∆CRISPR mutant) harbouring an aTc-inducible protospacer RNA. Arrowhead indicates addition of 100 ng ml–1 aTc. Each data point represents the mean ± s.e.m. of three biological replicates. b, Wild-type and ∆CRISPR L. seeligeri cultures carrying an aTc-inducible target transcript were exposed to 100 ng ml–1 aTc for 3 h as in a, then diluted (at time 0 h) to OD600 = 0.05 in fresh medium in the presence or absence of aTc, and growth was monitored over 24 h. Each curve represents the mean ± s.e.m. of three biological replicates. c, Immediate reduction in CFU upon phage infection of L. ivanovii RR3 or ΩCRISPRVI strains. The indicated strains were infected with ϕRR4 at an MOI of 2, and CFU titres in the infected cultures were monitored over time. Pre-infection (P) and mock-infection titres were also measured. Each bar represents the mean ± s.e.m. of three biological replicates. d, Cell vitality within ΩCRISPRVI cultures during phage infection. Cell vitality was measured in samples of cultures from c by monitoring conversion of nonfluorescent resazurin to fluorescent resorufin at each time point. The resorufin signal from heat-killed cells was subtracted from all samples as background, and each signal was normalized to the pre-infection value. Live cell standards (10% and 50%, mixed with heat-killed cells) are shown to demonstrate the quantitative capability of the vitality assay. Each bar represents the mean ± s.e.m. of three biological replicates. e, Phage-susceptible L. ivanovii ΩCRISPRVI(spcP) cells harbouring a spacer against an aTc-inducible plasmid target RNA (or empty vector control) were treated with aTc for 1 h to pre-activate Cas13a, then infected with ϕRR4 at an MOI of 1. Viable CFUs were counted before infection (PRE), 7 h after infection (T7) or after mock infection (UN). Two-sided Student’s t-test, ***P = 0.0005. Each bar represents the mean ± s.e.m. of three biological replicates.
Extended Data Fig. 6 Absence of CRISPR-resistant escape mutants and validation of engineered escaper phage.
a, Efficiency of plaque-forming assays with wild-type ϕRR4 and engineered spcA-escaper phage ϕRR4acr infecting L. ivanovii RR3 and ΩCRISPRVI(spcA) strains. Phages were diluted and spotted onto top agar lawns containing the indicated strain. Escaper plaques were not observed in the presence of type VI CRISPR targeting. The ϕRR4acr mutant, which lacks the acr region targeted by spcA, is viable and evades CRISPR targeting. Representative of two biological replicates. b, As in a, but testing the spcE-escaper phage ϕRR4early and the ΩCRISPRVI(spcE) strain. Representative of two biological replicates. c, Design of the ϕRR4acr mutant, harbouring a deletion of the putative anti-CRISPR genes of ϕRR4. d, Design of the ϕRR4early mutant, depicting the wild-type and mutant spcE target sequence. e, Cells pre-infected with wild-type ϕRR4 continue to adsorb escaper phage 2 h after infection. ΩCRISPRVI(spcE) cells were infected with wild-type ϕRR4 at an MOI of 5 for 2 h (an uninfected control is shown for comparison), then washed three times with fresh medium to remove free phages. ϕRR4early was added to cells (or to a cell-free control) at an MOI of 0.1 for 5 min, cells and bound phage were pelleted, and free phage in the supernatant were quantified as PFUs on a lawn of ΩCRISPRVI(spcE) cells. Mean PFU values are shown from two biological replicates. f, Efficiency of plaque-forming assay using RR3 and ΩCRISPRII(spcE) strains. We generated a ΩCRISPRII(spcE) strain carrying the type II-A CRISPR system from S. pyogenes programmed with spcE against ϕRR4. This strain has very limited immunity to wild-type ϕRR4, but is highly immune to the ϕRR4acr mutant, which lacks anti-CRISPR–Cas9 genes. Cas9-resistant ϕRR4acr escaper plaques are evident in the plaque assay (yellow arrowheads). One escaper, ϕRR4acr-esc, was isolated and confirmed to be resistant to Cas9 targeting. Representative of two biological replicates
Supplementary information
Supplementary Information 1
Expression and enrichment values for ϕRR4 spacers and corresponding target transcripts. Position, sequence, and enrichment data for all ϕRR4-targeting spacers detected at least 10 times in the pre-infection dataset. Spacer enrichment values are the normalized read counts 5h post-infection divided by those pre-infection. Expression of the corresponding protospacer sequence at 10, 20, 40, 60, and 90 minutes post-infection is also shown for each spacer (average RNA-seq read count per nucleotide across each protospacer at the indicated time point). Spacers are divided into anti-CRISPR (ACR), early lytic (EARLY), late lytic (LATE) and lysogeny (LYSOGENY) of the ϕRR4 genome.
41586_2019_1257_MOESM3_ESM.xlsx
Supplementary Information 2 L. ivanovii RR3 and ΩCRISPR(spcE) transcript cleavage ratios during ϕRR4 infection. Position, predicted transcriptional start site (TSS), and cleavage ratio for all L. ivanovii RR3 transcripts in wild-type RR3 and ΩCRISPR(spcE) 105 minutes post infection with ϕRR4 phage. TSS was predicted from 5’ end mapping data by determining the site containing the most 5’ end reads in the 500nt region preceding the gene/operon start codon. For detected transcripts (>20 reads in each sample) the cleavage ratio was calculated as the sum of reads downstream of the TSS (representing cleavage products) divided by the sum within (+/- 5 nt) the TSS (representing primary transcripts). The ratio of cleavage ratios in ΩCRISPR(spcE) to those in wt RR3 is also shown as a measure of CRISPR-dependent cleavage in each transcript.
41586_2019_1257_MOESM4_ESM.xlsx
Supplementary Information 3 L. ivanovii RR3 and ΩCRISPR(spcL) transcript cleavage ratios during ϕRR4 infection. Position, predicted transcriptional start site (TSS), and cleavage ratio for all L. ivanovii RR3 transcripts in wild-type RR3 and ΩCRISPR(spcL) 105 minutes post infection with ϕRR4 phage. TSS was predicted from 5’ end mapping data by determining the site containing the most 5’ end reads in the 500nt region preceding the gene/operon start codon. For detected transcripts (>20 reads in each sample) the cleavage ratio was calculated as the sum of reads downstream of the TSS (representing cleavage products) divided by the sum within (+/- 5 nt) the TSS (representing primary transcripts). The ratio of cleavage ratios in ΩCRISPR(spcL) to those in wt RR3 is also shown as a measure of CRISPR-dependent cleavage in each transcript.
41586_2019_1257_MOESM5_ESM.xlsx
Supplementary Information 4 L. seeligeri WT and ∆CRISPR transcript cleavage ratios during target transcription. Position, predicted transcriptional start site (TSS), and cleavage ratio for all L. seeligeri ATCC35967 transcripts in wild-type and ∆CRISPR carrying an aTc-inducible target transcript 15 minutes after the addition of 100 ng/mL aTc. TSS was predicted from 5’ end mapping data by determining the site containing the most 5’ end reads in the 500nt region preceding the gene/operon start codon. For detected transcripts (>20 reads in each sample) the cleavage ratio was calculated as the sum of reads downstream of the TSS (representing cleavage products) divided by the sum within (+/- 5 nt) the TSS (representing primary transcripts). The ratio of cleavage ratios in wild-type to those in ∆CRISPR is also shown as a measure of CRISPR-dependent cleavage in each transcript.
Supplementary Information 5
Tables of bacterial strains, bacteriophages, plasmids, oligonucleotides and synthetic DNA constructs used in this study.
Rights and permissions
About this article
Cite this article
Meeske, A.J., Nakandakari-Higa, S. & Marraffini, L.A. Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570, 241–245 (2019). https://doi.org/10.1038/s41586-019-1257-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1257-5
This article is cited by
-
Transcriptional dynamics during Rhodococcus erythropolis infection with phage WC1
BMC Microbiology (2024)
-
dCas13-mediated translational repression for accurate gene silencing in mammalian cells
Nature Communications (2024)
-
DNA-targeting short Argonautes complex with effector proteins for collateral nuclease activity and bacterial population immunity
Nature Microbiology (2024)
-
CRISPR/Cas genome editing in plants: mechanisms, applications, and overcoming bottlenecks
Functional & Integrative Genomics (2024)
-
CRISPR-Cas System: A New Dawn to Combat Antibiotic Resistance
BioDrugs (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.