Abstract
Pancreatic ductal adenocarcinoma (PDAC) has a dismal prognosis largely owing to inefficient diagnosis and tenacious drug resistance. Activation of pancreatic stellate cells (PSCs) and consequent development of dense stroma are prominent features accounting for this aggressive biology1,2. The reciprocal interplay between PSCs and pancreatic cancer cells (PCCs) not only enhances tumour progression and metastasis but also sustains their own activation, facilitating a vicious cycle to exacerbate tumorigenesis and drug resistance3,4,5,6,7. Furthermore, PSC activation occurs very early during PDAC tumorigenesis8,9,10, and activated PSCs comprise a substantial fraction of the tumour mass, providing a rich source of readily detectable factors. Therefore, we hypothesized that the communication between PSCs and PCCs could be an exploitable target to develop effective strategies for PDAC therapy and diagnosis. Here, starting with a systematic proteomic investigation of secreted disease mediators and underlying molecular mechanisms, we reveal that leukaemia inhibitory factor (LIF) is a key paracrine factor from activated PSCs acting on cancer cells. Both pharmacologic LIF blockade and genetic Lifr deletion markedly slow tumour progression and augment the efficacy of chemotherapy to prolong survival of PDAC mouse models, mainly by modulating cancer cell differentiation and epithelial–mesenchymal transition status. Moreover, in both mouse models and human PDAC, aberrant production of LIF in the pancreas is restricted to pathological conditions and correlates with PDAC pathogenesis, and changes in the levels of circulating LIF correlate well with tumour response to therapy. Collectively, these findings reveal a function of LIF in PDAC tumorigenesis, and suggest its translational potential as an attractive therapeutic target and circulating marker. Our studies underscore how a better understanding of cell–cell communication within the tumour microenvironment can suggest novel strategies for cancer therapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All source data for full immunoblot scan images and statistical analyses presented in figure panels are included in the Supplementary Information. RNA sequencing data have been deposited in the Gene Expression Omnibus under accession numbers GSE99187 and GSE119694. All raw mass spectrometry data have been deposited to the MassIVE repository (ftp://massive.ucsd.edu/MSV000081136).
Code availability
The custom scripts for quantification of immunohistochemistry images with Image J and multiplex immunofluorescence images with Imaris will be freely available upon reasonable request.
Change history
30 November 2021
A Correction to this paper has been published: https://doi.org/10.1038/s41586-021-04176-w
References
Chu, G. C., Kimmelman, A. C., Hezel, A. F. & DePinho, R. A. Stromal biology of pancreatic cancer. J. Cell. Biochem. 101, 887–907 (2007).
Feig, C. et al. The pancreas cancer microenvironment. Clin. Cancer Res. 18, 4266–4276 (2012).
Omary, M. B., Lugea, A., Lowe, A. W. & Pandol, S. J. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J. Clin. Invest. 117, 50–59 (2007).
Mahadevan, D. & Von Hoff, D. D. Tumor–stroma interactions in pancreatic ductal adenocarcinoma. Mol. Cancer Ther. 6, 1186–1197 (2007).
Erkan, M. et al. StellaTUM: current consensus and discussion on pancreatic stellate cell research. Gut 61, 172–178 (2012).
Apte, M. V., Wilson, J. S., Lugea, A. & Pandol, S. J. A starring role for stellate cells in the pancreatic cancer microenvironment. Gastroenterology 144, 1210–1219 (2013).
Tape, C. J. et al. Oncogenic KRAS regulates tumor cell signaling via stromal reciprocation. Cell 165, 910–920 (2016).
Corcoran, R. B. et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 71, 5020–5029 (2011).
Fukuda, A. et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 19, 441–455 (2011).
Lesina, M. et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 19, 456–469 (2011).
Sherman, M. H. et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 159, 80–93 (2014).
Nicola, N. A. & Babon, J. J. Leukemia inhibitory factor (LIF). Cytokine Growth Factor Rev. 26, 533–544 (2015).
Peñuelas, S. et al. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15, 315–327 (2009).
Liu, S. C. et al. Leukemia inhibitory factor promotes nasopharyngeal carcinoma progression and radioresistance. J. Clin. Invest. 123, 5269–5283 (2013).
Albrengues, J. et al. LIF mediates proinvasive activation of stromal fibroblasts in cancer. Cell Reports 7, 1664–1678 (2014).
Inda, M. M. et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 24, 1731–1745 (2010).
Öhlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Bressy, C. et al. LIF drives neural remodeling in pancreatic cancer and offers a new candidate biomarker. Cancer Res. 78, 909–921 (2018).
Biffi, G. et al. IL-1-induced JAK/STAT signaling is antagonized by TGFβ to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 9, 282–301 (2018).
Laklai, H. et al. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 22, 497–505 (2016).
Bayne, L. J. et al. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell 21, 822–835 (2012).
Pylayeva-Gupta, Y., Lee, K. E., Hajdu, C. H., Miller, G. & Bar-Sagi, D. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21, 836–847 (2012).
Kim, K. J., Alphonso, M., Schmelzer, C. H. & Lowe, D. Detection of human leukemia inhibitory factor by monoclonal antibody based ELISA. J. Immunol. Methods 156, 9–17 (1992).
Li, C. et al. Identification of pancreatic cancer stem cells. Cancer Res. 67, 1030–1037 (2007).
Hermann, P. C. et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 1, 313–323 (2007).
Kim, M. P. et al. ALDH activity selectively defines an enhanced tumor-initiating cell population relative to CD133 expression in human pancreatic adenocarcinoma. PLoS One 6, e20636 (2011).
Li, C. et al. c-Met is a marker of pancreatic cancer stem cells and therapeutic target. Gastroenterology 141, 2218–2227 (2011).
Fox, R. G. et al. Image-based detection and targeting of therapy resistance in pancreatic adenocarcinoma. Nature 534, 407–411 (2016).
Singh, A. & Settleman, J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 (2010).
Jameson, G. S. et al. A phase Ib/II pilot trial with nab-paclitaxel plus gemcitabine plus cisplatin in patients (pts) with stage IV pancreatic cancer. J. Clin. Oncol. 35, 341 (2017).
Ouyang, H. et al. Immortal human pancreatic duct epithelial cell lines with near normal genotype and phenotype. Am. J. Pathol. 157, 1623–1631 (2000).
Shackelford, D. B. et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell 23, 143–158 (2013).
Andrews, S. FastQC: a quality control tool for high throughput sequence data http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Babraham Bioinformatics, 2010).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Dauer, D. J. et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene 24, 3397–3408 (2005).
Rissin, D. M. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010).
Villén, J. & Gygi, S. P. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protocols 3, 1630–1638 (2008).
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Tian, R. et al. Combinatorial proteomic analysis of intercellular signaling applied to the CD28 T-cell costimulatory receptor. Proc. Natl Acad. Sci. USA 112, E1594–E1603 (2015).
Zheng, Y. et al. Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 499, 166–171 (2013).
Chen, W. et al. 3D-SISPROT: A simple and integrated spintip-based protein digestion and three-dimensional peptide fractionation technology for deep proteome profiling. J. Chromatogr. A 1498, 207–214 (2017).
Zhang, H., Li, X. J., Martin, D. B. & Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 (2003).
Peterson, A. C., Russell, J. D., Bailey, D. J., Westphall, M. S. & Coon, J. J. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol. Cell. Proteomics 11, 1475–1488 (2012).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Linkert, M. et al. Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010).
Roy, N. et al. PDX1 dynamically regulates pancreatic ductal adenocarcinoma initiation and maintenance. Genes Dev. 30, 2669–2683 (2016).
Robin, X. et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77 (2011).
Acknowledgements
We thank F. McCormick and M. Wang (UCSF) for sharing reagents and results, R. Shaw (Salk) for KPf/fL mice, M. Karin (UCSD) for mouse cancer samples and comments on the manuscript, H. Han and E. Menashi (Tgen) for experimental suggestions and reagents, J. Zimmerman and S. Simon for laboratory support, N. Cernac and J. Chang for laboratory assistance, J. Chambers and K. Suter-Brady for animal work, T. Zhang and K. Mcintyre for histology services. This work was supported by the following grants: Ministry of Science and Technology of China (2016YFA0501403) and National Natural Science Foundation of China (21575057) to R.T.; NIH CA014195, CA082683, CA080100, Lustgarten Foundation (awards 388246 and 552873), Helmsley Charitable Trust (2012-PG-MED002), and William Isacoff Research Foundation to T.H.; SU2C Pancreatic Cancer Dream Team (SU2C-AACR-DT-05-09, SU2C-AACR-DT-20-16) to D.D.V.H., R.M.E., G.W., T.H., A.M.L., T.R. and M.D.; NRSA F31CA206416 and T32GM007752 to N.K.L.; F32CA217033 to G.L.; Flinn Foundation to E.B.; NIH CA178015, CA227807 and CA222862 to E.A.C.; NIH CA197699 and CA186043 to T.R.; NIH CA155620 to A.M.L.; NIH P50CA127297, U01CA210240, P30CA36727 and 5R50CA211462 to P.M.G. and M.A.H. R.M.E. is a Howard Hughes Medical Institute Investigator and March of Dimes Chair in Molecular and Developmental Biology, and supported by Lustgarten, Ipsen/Biomeasure, Freeberg and William Isacoff Research Foundation. We thank Salk core facilities and staff, financially supported by NCI CCSG CA014195, including Advanced Biophotonics, Flow Cytometry, Functional Genomics, Integrative Genomics and Bioinformatics, Next Generation Sequencing and Transgenic Cores. Y.S. received a fellowship from the Helmsley Charitable Trust. T.H. is a Frank and Else Schilling American Cancer Society Professor and the Renato Dulbecco Chair in Cancer Research.
Reviewer information
Nature thanks Claus Jørgensen, Amina Ann Qutub and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Y.S. conceived the project, planned the research, performed or participated in all experiments and wrote the paper. W.G. performed mass spectrometry assays and helped prepare figures. N.K.L. and T.R. performed and analysed tumour-initiating cell-related data. P.H. and X.Y. performed PRM-MS. A.M.D. and T.R.D. provided human pancreatic tissues and FFPE specimens, and performed correlation analysis. M.R.-S. and K.E.D. provided reagents and participated in some mouse studies. C.E.A., G.L. and A.R.A. provided PSCs and organoid lines. G.E. and H.S. performed RNA-seq data analysis. J.M., E.T. and G.W. provided laboratory and animal work support. L.F., T.P.S. and U.M. developed image quantification algorithms. R.X. provided cancer tissue specimens and pathology consultation. D.D.V.H., C.R.B, E.B, P.M.G. and M.A.H. provided human PDAC serum specimens. M.L., S.E.U. and E.A.C. performed pathological grading. M.S. provided advice for fMRI. A.M.L. provided human serum specimens, pathological evaluation and manuscript comments. M.D., R.M.E., and G.M.W. provided intellectual support and manuscript comments. T.P. participated in the initial project conception. R.T. conceived the mass spectrometry screening, designed and participated in all mass spectrometry assays and wrote the paper. T.H. conceived and oversaw the project and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
E.B. is a scientific advisor to Fujifilm, Ipsen and Corcept. D.D.V.H. is an employee of McKesson, a co-founder of Stromatis Pharma and Systems Oncology, and a shareholder of McKesson, Medtronic, CerRx, SynDevRx, UnitedHealthare, Anthem, Capella Therapeutics and Cell Therapeutics. D.D.V.H. is a scientific advisor to DNAtrix, Esperance, Five Prime, Imaging Endpoints, Immoduon, Insys, Medical Prognosis Institute, miRNA Therapeutics, Senhwa Biosciences, Tolero, Trovagene, Alpha Cancer Techologies, Arvinas, Bellicum, CanBas, Horizon Discovery, Innate, Lixte Biotechnology, Oncolyze, RenovoRx, TD2, AADI, Aptose Biosciences, BiolineRx, CV6, CytomX, EMD Serono, Evelo, Fujifilm, Histogen, Intezyne, Kalos, Kura, Phosplatin, SOTIO, Strategia, Sun Biopharma, Synergene, Systems Imagination, 7 Hills, Actinium, Aduro Biotech, ARMO BioSciences, Cancer Prevention Pharmaceuticals, Defined Health, Geistlich Pharma, HUYA Bioscience, Immunophotonics, Novocure, ARIAD, Boston Biomedical, Corrona, Forma, Genzada, L.E.A.F., Oncology Venture, Reflexion Medical, TP Therapeutics, Verily, Athenex, Fate, FibroGen, Jounce Therapeutics, Samus Therapeutics, Sumitomo Dainippon, Aeglea Biotherapeutics, 2X Oncology, Innokeys, Novita, NuCana BioMed, Araxes, Ipsen, SciClone, TargaGenix, Trans Med, Veana Therapeutics, Biospecifics Technologies, Riptide Bioscience, Vicus Therapeutics, Codiak Biosciences, Decoy Biosystems, OSI, Agenus, Globe Life Science, Kelun–Klus Pharma, RadImmune, Samumed, SOBI, Adicet Bio, BioXCel Therapeutics, Bryologyx, Celgene, Helix BioPharma, Sirnoamics, AiMed Bio, Boston Scientific, Corcept, Erimos and Gimbal.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Combinatorial mass spectrometry analyses to characterize the paracrine communication between PSCs and PCCs.
a, Phosphotyrosine proteomic analysis of intracellular signalling changes in MIA PaCa2 cells in response to stimulation with hPSC-conditioned medium. n = 2 biological replicates. b, Summary of phosphotyrosine proteomic analysis data in PANC1 and MIA PaCa2 cells in response to stimulation with hPSC-conditioned medium. c, Workflow of the analysis of secretome proteomic assays. Proteins identified with at least three spectral counts were counted, and only those identified in both biological replicates were considered. Proteins uniquely secreted by each cell type were defined as those with more than tenfold differences in spectral count. n = 2 biological replicates. d, DAVID gene ontology (GO) analysis of the protein sets uniquely secreted by MIA PaCa2 cells and hPSC identified the top ten enriched GO terms of molecular function for each cell type. e, Pearson correlation analysis to validate the quantification reproducibility of STAT3 IP–MS assays by label-free quantification (LFQ) between biological replicates. n = 3 biological replicates for both control and stimulation with PSC-conditioned medium.
Extended Data Fig. 2 Dysregulated LIF is a key driver for STAT3 activation in PDACs.
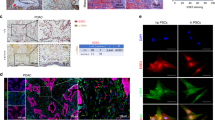
a, Immunoblot analyses of the response of various human PCC and PSC cell lines to LIF stimulation using STAT3 phosphorylation at Y705 (pSTAT3) as a readout. b, Immunoblot analyses of LIF-stimulated downstream intracellular signalling changes in three representative PCC lines, KP4, MIA PaCa2 and PANC1, over a 2-h time course. Stimulation with recombinant human epidermal growth factor (EGF) 10 ng ml−1 for 10 min was used as positive control for pAKT1 and pERK1/2 activation. c, d, Immunoblot analyses of pSTAT3 in MIA PaCa2 and PANC1 cells when LIF signalling was blocked by either shRNA knockdown of LIFR (c) or immune-inactivation of LIF using anti-LIF monoclonal antibody (d). For LIF stimulation (a–d), 1 ng ml−1 recombinant human LIF was applied for 15 min, and at least three independent experiments were performed; representative images are presented. e, LIFR expression in PCCs by qPCR and its positive correlation with the response intensity to LIF as illustrated in the heat map. n = 2 biological replicates. f, LIF secretion by PCCs and PSCs quantified by ELISA, and its negative correlation with the corresponding response intensity to LIF. n = 2 biological replicates. g, Cellular localization of Lif mRNA in pancreatic tumour tissues from KPf/fCL mice was examined by multiplex fluorescent RNAscope assays. Ptprc (also known as Cd45) mRNA was co-stained to mark immune cells, and KRT19 was stained by immunofluorescence to mark cancer cells. Scale bars: red, 200 µm; white, 50 µm. h–j, pSTAT3 analysed by immunoblotting (h) and immunohistochemistry (i) in mouse pancreatic tissues, and by immunohistochemistry in human pancreatic tissues during the pathogenesis of PDAC (j). Scale bars: black, 500 µm; blue, 100 µm. For the histology assays on tissue sections (g, i, j), at least three tumour samples were stained and analysed, and representative images are presented.
Extended Data Fig. 3 Intrinsic LIFR signalling in PCCs affects pancreatic cancer progression but not initiation and acinar-ductal metaplasia development.
a, Cellular localization of Lifr mRNA in mouse pancreatic tissues by RNAscope assays. Postn mRNA was stained to mark the stromal PSC cells. b, Schematic illustration of Lifr mutant allele structure and conversion into conditional deletion mutant allele by FLPo excision. c, d, Characterization of primary mouse embryonic fibroblasts isolated from Lifrf/f and LIFRf/+ mice, respectively, by PCR analysis to determine the genotype of the Lifr allele and loxP-flanked exon deletion by adenovirus-introduced Cre (c), and by immunoblot analysis to validate the knockout of LIFR proteins and loss of response to LIF (d). +, wildtype allele; f, loxP-flanked mutant allele; ∆, mutant allele with the loxP-flanked exon deleted. e, Representative histological images characterizing tumours from LifrWTKPf/fCL or Lifrf/fKPf/fCL mice at the endpoint of the survival study presented in Fig. 2b. f, g, Histology characterization of tumour development in LifrWTKPf/fCL or Lifrf/fKPf/fCL mice at 3, 5 and 7 weeks of age (f), and quantification of cancer cell abundance (g); n = 6 mice per condition. h, i, Pathological grade of tumour stage (h) and histological quantification of cancer and stromal PSC cell abundance (i) for pancreatic tissues collected from LifrWTKC or Lifrf/fKC mice treated with 250 mg kg−1 caerulein by daily intraperitoneal injection, starting at seven weeks of age, for five days and rested for five days to allow tumour development. ADM, acinar-ductal metaplasia. j, Histology characterization of caerulein-treated LifrWT;Pdx1-Cre or Lifrf/f;Pdx1-Cre mice by Masson’s trichrome stain. Littermates at seven weeks of age were subjected to daily 250 mg kg−1 body weight caerulein treatment by intraperitoneal injection for seven days, and pancreas tissues were collected one or seven days after the last injection to examine the acinar-ductal metaplasia formation and resolution. No differences were noticed between two genotypes. n = 5 mice per condition. k, RNA in situ hybridization by BaseScope assays using probes specifically targeting the loxP-flanked exon 4 of Lifr to examine the escaper cancer cells still maintaining Lifr expression owing to incomplete deletion. Scale bars: black, 1,000 µm; blue, 200 µm; yellow, 50 µm. Statistical significance was determined by two-way ANOVA (h) or two-tailed unpaired Student’s t-test.
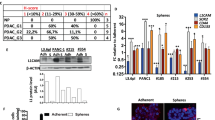
Extended Data Fig. 4 LIFR signalling in PCCs modulates cancer cell differentiation.
a, b, Flow cytometry analysis of the total cell counts (a) and relative frequency (b) of tumour-initiating cell populations in individual tumours. c, Functional evaluation of tumour-initiating cell abundance by in vitro sphere formation. d–i, Heat maps showing differential gene expression comparing the EpCAM+ cancer cells purified from individual tumours of LifrWTKPf/fCL or Lifrf/fKPf/fCL mice. Colours correspond to standardized expression of genes. n = 4 mice per treatment. d, Hierarchical clustering of all the 1,129 differentially expressed genes (FDR <0.05, log2(fold change) >0.8 and FPKM >2 in at least four samples). Heat maps of genes related to tumour-initiating cell markers (e), STAT3 downstream targets (f), and Gem response (g). h, i, GSEA. j, l, Expression of Csf2 (encoding GM-CSF) and Ccl11 in EpCAM+ PCCs (n = 4), PDGFRα+ cancer-associated fibroblasts (CAFs; n = 3) and CD45+ tumour infiltrating lymphocytes (TILs; n = 3), purified by FACS from tumours of KPf/fCL mice, determined by RNA-seq analysis. k, m, n, Multiplex ELISA analysis of GM-CSF and CCL11 levels in normal, caerulein-induced chronic pancreatitis and PDAC tissues of KPf/fCL mice showed that the increase in GM-CSF was induced only when tumours developed (k), whereas the increase in CCL1L level was induced in both chronic pancreatitis and PDAC consistently, both in mouse models (m) and human disease (n), supporting the notion that CCL11 is a cytokine specifically produced by PSCs, whereas GM-CSF is produced specifically by PCCs. o, Cellular localization of Ccl11 mRNA in pancreatic cancer tissues from KPf/fCL mice was examined by multiplex fluorescent RNAscope assays. Ptprc mRNA was co-stained to mark immune cells, and KRT19 was stained by immunofluorescence to mark cancer cells. n = 3 tumours. Scale bars: yellow, 500 µm; white, 100 µm.
Extended Data Fig. 5 Preclinical study in KPf/fCL mouse model reveals therapeutic beneficial effects of LIF blockade by anti-LIF monoclonal antibody.
a–d, Anti-LIF monoclonal antibody production and characterization. a, Silver staining of control mouse IgG and purified anti-LIF monoclonal antibody used for preclinical treatment. b, c, Immunoblot analysis to evaluate the activity and specificity of anti-LIF monoclonal antibody against various forms of LIF (rhLIF, recombinant human LIF; ehLIF, eukaryotically secreted human LIF; rmLIF, recombinant mouse LIF) and IL-6 (b), revealing that the LIF monoclonal antibody has weaker neutralizing activity against mLIF compared to against hLIF (c). d, Pilot test of anti-LIF monoclonal antibody in KPf/fCL mice for dosage optimization. KPf/fCL mice at 42 days of age were administered 25 mg per kg (body weight) of LIF monoclonal antibody by three intraperitoneal injections on alternate days, and one day after the last injection tumours were collected for histology analysis. Adjacent sections were used for the indicated staining. Representative images from three independent experiments or mice are shown. e, Regimen for the preclinical therapeutic treatment. KPf/fCL mice at five weeks of age were randomly enrolled into four cohorts. In phase one, during the first 12 days, 25 mg per kg (body weight) anti-LIF monoclonal antibody or control IgG were administered by intraperitoneal injection, together with 50 mg kg−1 Gem or vehicle at standard dosage every third day for four administrations. This was followed by weekly cycles as phase two with antibodies three times and Gem two times weekly. f, Histological characterization with representative images and cell abundance quantification. g, Relative abundance of EpCAM+ cancer cells quantified by flow cytometry analysis. h, Double immunofluorescence staining to confirm the cell type-specific expression of cytosolic protein KRT19 and nuclear protein PDX1 as the PCC marker. n = 3 tumour tissues. i, Double immunofluorescence staining of nucleus-localized proteins Ki67 (used as a proliferation marker) and PDX1 (as the PCC marker), and quantification of proliferating cancer cell frequency as the fraction of proliferating cancer cells (Ki67+PDX1+DAPI+) over total cancer cells (PDX1+DAPI+). j, Cleaved Caspase 3 immunohistochemistry analysis to assess apoptosis. Scale bars: yellow, 300 µm; black, 100 µm; white, 50 µm. Statistical significance was determined by two-tailed unpaired Student’s t-test.
Extended Data Fig. 6 LIF blockade alleviates chemoresistance directly by affecting cancer cell differentiation.
a, Representative histological images showing different tumour differentiation status. b–d, Flow cytometry analysis of the total cell count (b) and relative frequency (c) of tumour-initiating cell populations in individual tumours, and the gating strategy exemplified with representative contour plots (d). e, Multiplex immunofluorescence staining of nuclear proteins ZEB1 (as a mesenchymal cell marker) and PDX1 (as the PCC marker) and quantification of mesenchymal cancer cell frequency as the fraction of ZEB1+PDX1+DAPI+ cancer cells over PDX1+DAPI+ total cancer cells. f, Sphere formation assays on primary PCCs with or without Lifr deficiency in response to low concentration (3 nM) Gem treatment. n = 9 per condition. g–i, Histological characterization of tumour tissues from the maintenance study (in Fig. 4c–e) and representative images of AB-PAS staining for well-differentiated cancer cells with acidic and neutral mucin stained in blue and magenta, respectively (g), multiplex immunofluorescence of ZEB1 and PDX1 proteins for the mesenchymal cancer cell frequency quantification and EMT (h), and of cleaved caspase-3 immunohistochemistry analysis for apoptosis (i). Scale bars: black, white, 300 µm; blue, 100 µm; yellow, 50 µm. Statistical significance was determined by two-tailed unpaired Student’s t-test.
Extended Data Fig. 7 LIF blockade profoundly affects gene expression in cancer cells analysed by RNA-seq.
a–e, Differential gene expression comparing the EpCAM+ cancer cells purified from individual tumours of KPf/fCL mice treated with either chemo (GI) or combo (GA) is shown in heat maps. Colours correspond to standardized expression of genes. a, Hierarchical clustering of all 1,624 differentially expressed genes (false discovery rate, FDR <0.05, log2(fold change) >0.8 and FPKM >2 in at least four samples). b, GSEA. Heat maps presenting the expression levels of genes related to tumour-initiating cell markers (c), STAT3 downstream targets (d) and Gem response (e). f, Venn diagram comparing the differentially expressed genes by pharmacologic LIF blockade and genetic Lifr deficiency revealed 811 consensus genes. g, Hierarchical clustering of 811 consensus genes with differential expression by pharmacologic LIF blockade and genetic Lifr deficiency.
Extended Data Fig. 8 Correlation analysis between tissue LIF levels and clinical parameters of PDAC.
a–c, ELISA of LIF levels (a), and immunoblot and immunohistochemistry analyses of STAT3 activation in mouse pancreatic tissues at various stages of PDAC pathogenesis. Scale bars: black, 200 µm; blue, 50 µm. d, Mouse serum LIF levels at various stages of pancreas pathogenesis analysed by ELISA. e, Summary table of clinical parameters of the human cases from whom pancreatic tissue samples were collected for ELISA assays and correlation analysis. f, Correlation analysis of human tissue LIF levels and indicated clinical parameters by χ2-analysis. g, Correlation analysis between tissue LIF mRNA levels and disease-free survival of patients with stage I or IIa PDAC at diagnosis by Mantel–Cox log-rank test and presented as a Kaplan–Meier survival curve. Data derived from the TCGA database. h, Correlation analysis between the changes in circulating LIF or CA19-9 levels and tumour status scored by RECIST grade during multi-cycle therapeutic treatment by ROC analysis. n = 51 data points. i, Serum LIF levels in patients with various solid cancers by Simoa ELISA. j, Graphic summary of LIF action in pancreatic carcinogenesis.
Extended Data Fig. 9 PRM-MS assay development and analysis.
a, Schematic workflow of the glycoprotein enrichment strategy followed with PRM-MS assays. b, Identified unique peptides of the targeted proteins in human pancreatic tissues. c, Reproducibility evaluation of the tissue sample preparation and PRM-MS analysis. d, Summary table of clinical parameters of the human cases from whom pancreatic tissue samples were collected for PRM-MS analysis. e, f, LIF protein levels in human primary tumour and paired normal tissue samples from patients with pancreatic cancer (e) and patients with various solid cancers (f), quantified by PRM-MS assays. g, h, LIFR and GP130 protein levels in human pancreatic tissues quantified by PRM-MS assays. Statistical significance was determined by two-tailed unpaired Student’s t-test (f) or one-way ANOVA (e, g, h).
Extended Data Fig. 10 Comparison of LIF and IL-6 in PDACs.
a, b, MS and ELISA quantification of IL-6 family cytokines in hPSC-conditioned medium. n = 2 biological replicates. c–e, ELISA quantification of IL-6 (c) and IL-11 (d) in conditioned media from various human PCC lines, and LIF and IL-6 in conditioned media from various hPSC or normal fibroblast cells (e). n = 2 biological replicates. f–i, ELISA quantification of LIF and IL-6 levels in mouse and human pancreatic tissues (f, h) and sera (g, i), respectively. j, k, Correlation analysis between human tissue IL-6 levels and tumour differentiation status by Kruskal–Wallis test (j) or overall survival by Pearson correlation test (k). l–n, Correlation analysis between tissue IL6 mRNA levels and overall survival by Mantel–Cox log-rank test presented as Kaplan–Meier survival curve (l), and mRNA expression compared by Wilcoxon matched-pairs rank test and presented as Tukey box-and-whisker plots (m, n); RNA-seq data of 170 PDAC tissues were derived from the TCGA database. o, Cellular localization of mRNA expression in human pancreatic tissues examined by RNAscope assays. KRT19 or POSTN were co-stained to mark PCC and PSC cells respectively. p, q, Immunoblot analysis of pSTAT3 activation in indicated human pancreatic cancer cell lines in response to stimulation with 1 ng ml−1 recombinant human IL-6 or LIF (p) or by hPSC-conditioned medium with or without immune inactivation of LIF and/or IL-6 using neutralizing antibodies (q). All cells were stimulated for 15 min. Representative images from three independent experiments are shown.
Supplementary information
Supplementary Information
This file contains a Supplementary Discussion, the uncropped blots (Supplementary Figure 1) and a list of antibodies used in the study (Supplementary Table 1).
Source data
Rights and permissions
About this article
Cite this article
Shi, Y., Gao, W., Lytle, N.K. et al. Targeting LIF-mediated paracrine interaction for pancreatic cancer therapy and monitoring. Nature 569, 131–135 (2019). https://doi.org/10.1038/s41586-019-1130-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1130-6
This article is cited by
-
Tumor immune microenvironment-based therapies in pancreatic ductal adenocarcinoma: time to update the concept
Journal of Experimental & Clinical Cancer Research (2024)
-
Combination of bazedoxifene with chemotherapy and SMAC-mimetics for the treatment of colorectal cancer
Cell Death & Disease (2024)
-
DeKinomics pulse-chases kinase functions in living cells
Nature Chemical Biology (2024)
-
Leukemia inhibitory factor suppresses hepatic de novo lipogenesis and induces cachexia in mice
Nature Communications (2024)
-
CACNA2D1 regulates the progression and influences the microenvironment of colon cancer
Journal of Gastroenterology (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.