Abstract
The HIV-1 envelope glycoprotein (Env) trimer mediates cell entry and is conformationally dynamic1,2,3,4,5,6,7,8. Imaging by single-molecule fluorescence resonance energy transfer (smFRET) has revealed that, on the surface of intact virions, mature pre-fusion Env transitions from a pre-triggered conformation (state 1) through a default intermediate conformation (state 2) to a conformation in which it is bound to three CD4 receptor molecules (state 3)8,9,10. It is currently unclear how these states relate to known structures. Breakthroughs in the structural characterization of the HIV-1 Env trimer have previously been achieved by generating soluble and proteolytically cleaved trimers of gp140 Env that are stabilized by a disulfide bond, an isoleucine-to-proline substitution at residue 559 and a truncation at residue 664 (SOSIP.664 trimers)5,11,12,13,14,15,16,17,18. Cryo-electron microscopy studies have been performed with C-terminally truncated Env of the HIV-1JR-FL strain in complex with the antibody PGT15119. Both approaches have revealed similar structures for Env. Although these structures have been presumed to represent the pre-triggered state 1 of HIV-1 Env, this hypothesis has never directly been tested. Here we use smFRET to compare the conformational states of Env trimers used for structural studies with native Env on intact virus. We find that the constructs upon which extant high-resolution structures are based predominantly occupy downstream conformations that represent states 2 and 3. Therefore, the structure of the pre-triggered state-1 conformation of viral Env that has been identified by smFRET and that is preferentially stabilized by many broadly neutralizing antibodies—and thus of interest for the design of immunogens—remains unknown.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Code availability
The full source code of SPARTAN34, which was used for all analysis of smFRET data, is available at http://www.scottcblanchardlab.com/software.
References
Harrison, S. C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280, 1884–1888 (1998).
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998).
Liu, J., Bartesaghi, A., Borgnia, M. J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008).
Ozorowski, G. et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363 (2017).
Haynes, B. F. & Burton, D. R. Developing an HIV vaccine. Science 355, 1129–1130 (2017).
Sanders, R. W. & Moore, J. P. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol. Rev. 275, 161–182 (2017).
Munro, J. B. et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763 (2014).
Herschhorn, A. et al. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelopeglycoproteins. m Bio 7, e01598-e16 (2016).
Ma, X. et al. HIV-1 Env trimer opens through an asymmetric intermediate in which individual protomers adopt distinct conformations. eLife 7, e34271 (2018).
Sanders, R. W. et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618 (2013).
Julien, J. P. et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483 (2013).
Lyumkis, D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490 (2013).
Pancera, M. et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461 (2014).
Wang, H. et al. Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop. Proc. Natl Acad. Sci. USA 113, E7151–E7158 (2016).
Bartesaghi, A., Merk, A., Borgnia, M. J., Milne, J. L. & Subramaniam, S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Biol. 20, 1352–1357 (2013).
Gristick, H. B. et al. Natively glycosylated HIV-1 Env structure reveals new mode for antibody recognition of the CD4-binding site. Nat. Struct. Mol. Biol. 23, 906–915 (2016).
Kwon, Y. D. et al. Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531 (2015).
Lee, J. H., Ozorowski, G. & Ward, A. B. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048 (2016).
Chuang, G. Y. et al. Structure-based design of a soluble prefusion-closed HIV-1 Env trimer with reduced CD4 affinity and improved immunogenicity. J. Virol. 91, e02268-e16 (2017).
Sakin, V. et al. A versatile tool for live-cell imaging and super-resolution nanoscopy studies of HIV-1 Env distribution and mobility. Cell Chem. Biol. 24, 635–645 (2017).
Sen, J., Jacobs, A. & Caffrey, M. Role of the HIV gp120 conserved domain 5 in processing and viral entry. Biochemistry 47, 7788–7795 (2008).
Alsahafi, N., Debbeche, O., Sodroski, J. & Finzi, A. Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS ONE 10, e0122111 (2015); correction 10, e0129405 (2015).
Pancera, M. et al. Crystal structures of trimeric HIV envelope with entry inhibitors BMS-378806 and BMS-626529. Nat. Chem. Biol. 13, 1115–1122 (2017).
Caskey, M. et al. Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117. Nature 522, 487–491 (2015).
Caskey, M. et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 23, 185–191 (2017).
Nishimura, Y. et al. Early antibody therapy can induce long-lasting immunity to SHIV. Nature 543, 559–563 (2017).
Sok, D. et al. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature 548, 108–111 (2017).
Gardner, M. R. et al. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature 519, 87–91 (2015).
Castillo-Menendez, L. R., Nguyen, H. T. & Sodroski, J. Conformational differences between functional human immunodeficiency virus envelope glycoprotein trimers and stabilized soluble trimers. J. Virol. 93, e01709–e01718 (2019).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Lin, C. W. & Ting, A. Y. Transglutaminase-catalyzed site-specific conjugation of small-molecule probes to proteins in vitro and on the surface of living cells. J. Am. Chem. Soc. 128, 4542–4543 (2006).
Yin, J., Lin, A. J., Golan, D. E. & Walsh, C. T. Site-specific protein labeling by Sfp phosphopantetheinyl transferase. Nat. Protoc. 1, 280–285 (2006).
Juette, M. F. et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344 (2016).
Dave, R., Terry, D. S., Munro, J. B. & Blanchard, S. C. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys. J. 96, 2371–2381 (2009).
Aitken, C. E., Marshall, R. A. & Puglisi, J. D. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J. 94, 1826–1835 (2008).
McKinney, S. A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Zhong, P. et al. Cell-to-cell transmission can overcome multiple donor and target cell barriers imposed on cell-free HIV. PLoS ONE 8, e53138 (2013).
Poss, M. & Overbaugh, J. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73, 5255–5264 (1999).
Acknowledgements
We thank A. B. Ward, R. W. Sanders and P. J. Bjorkman for discussions, R. Blakemore for assistance with molecular modelling, D. Burton and M. Feinberg for reagents including PGT121, PGT122, PGT145, PGT151 and NC-Cow1, NC-Cow8, NC-Cow9 and NC-Cow10 antibodies, and the AIDS Research and the Reference Reagent Program (Division of AIDS, NIAID, NIH) for the antibodies 3BNC117, 10-1074, PG9 and PG16. This work was supported by NIH grants RO1 GM116654 and UM1 AI100645 to W.M., RO1 GM098859 to S.C.B., RO1 AI124982 and RO1 AI100645 to J.G.S., K22 AI116262 to J.B.M., CRC Tier 2 RCHS0235 and a CIHR foundation grant 352417 to A.F., PO1 GM056550 to W.M., J.G.S., S.C.B, A.B.S. and C.A., by a Brown Coxe Fellowship to M.L., a fellowship from the China Scholarship Council-Yale World Scholars to X.M., by the International AIDS Vaccine Initiative’s (IAVI’s) Neutralizing Antibody Consortium to P.D.K. and by the Intramural Research Program of the Vaccine Research Center (NIAID, NIH) to P.D.K. and A.B.M., and by the SFB1129 and the Emmy-Noether programme (project number 317530061) of the German Research Foundation to E.A.L. and I.N.-S., respectively.
Reviewer information
Nature thanks David Millar, Alexandra Trkola and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.L., X.M., L.R.C.-M., N.A., J.G., J.B.M., A.F., P.D.K., S.C.B., J.G.S. and W.M. conceived these experiments. M.L., X.M., U.E., N.R., K.W., J.R.G., and B.P.C. performed mutagenesis, and M.L. and X.M. generated fluorescently labelled viruses. M.L., X.M. and D.S.T. performed labelling and smFRET imaging. M.L., X.M., S.C.B. and W.M. analysed the data. L.R.C.-M., J.G., M.C., N.A., A.F., D.P., B.Z., T.Z. and A.B.M. performed protein expression, purification and antibody-binding experiments. L.R.C.-M. performed negative-stain electron microscopy. J.B.M. and C.A. performed photophysical measurements and molecular dynamics simulations. A.S., J.A., A.B.S.III, I.N.-S., E.A.L., M.R.G. and M.F. provided reagents. M.L., P.D.K, S.C.B., J.G.S. and W.M. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.C.B. holds an equity interest in Lumidyne Technologies. M.F. is a co-founder of Emmune, a company developing eCD4-Ig for clinical use. E.A.L. holds the patent WO 2012/104422 A1. S.C.B., J.B.M. and W.M. hold the patent US 959385324 B2.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Tagged BG505 sgp140 SOSIP.664 proteins largely retain their known immunogenic features and preferentially sample state-2-like conformations.
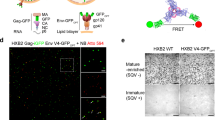
a, Schematics for wild-type (WT) BG505 Env and BG505 sgp140 SOSIP.664 with D7324 affinity tag; V1–Q3 peptide in green, V4–A1 peptide in red. b, Validation of tagged BG505 sgp140 SOSIP.664. Top, antigenic profile of 100% untagged (WT), 100% double-tagged V1V4 (V1–Q3 V4–A1), and 20:1 of untagged to double-tagged BG505 sgp140 SOSIP.664. Binding by the indicated VRC01, 17b, PG9, 19b, PGT151 and 902090 antibodies was assessed from two independent ELISA assays in hexaplets and displayed as percentage of 2G12 binding (mean ± s.d.). The epitope for the antibody 902090 was more exposed in the 100%-tagged BG505 sgp140 SOSIP.664 than in the untagged BG505 sgp140 SOSIP.664, although this was not the case for the 1:20 tagged:wild-type trimers used for our smFRET analyses. The insertion of the Q3 tag into all three V1 regions of Env may exert local effects on the V2 β-barrel that contains the 902090 epitope (residues 171–177). Bottom, reference-free negative-staining electron microscopy two-dimensional class averages with representative trimeric density map of the BG505 sgp140 SOSIP.664 (wild type:V1V4-tagged at a 20:1 ratio) used for smFRET imaging. A Fourier shell correlation is also provided. c, Antigenic characteristics of BG505 sgp140 DS-SOSIP.664 (left) and 100% V1V4-tagged BG505 DS-SOSIP.Mut4 (right), determined by MSD. Antibodies are labelled. CD4bs, CD4 binding site; CD4i, CD4-induced; V1V2, V1V2-directed; V3, V3 glycan site-directed; gp120/gp41, interface between gp120 and gp41. Antigenic profiles of BG505 DS-SOSIP.664 (left) and 100% V1V4-tagged DS-SOSIP.Mut4 (right) after V3-negative selection were assessed by a panel of CD4-induced antibodies (17 and 48b, with and without sCD4), CD4 binding site antibodies (VRC01, VRC03, b12 and weakly neutralizing F105), V1V2-directed antibodies (PGT145 and VRC26.25), V3 glycan site-directed antibodies (2G12, PGT121, PGT128) and weakly neutralizing V3-directed antibodies (447-52D, 3074 and 2557, with and without soluble CD4), gp41–gp120 interface antibodies (PGT151, 35O22, 8ANC195 and VRC34.01) and the negative-control antibody CR9114 (an influenza virus antibody that does not recognize HIV-1 Env). ECL, electrochemiluminescence. d, The indicated BG505 sgp140 SOSIP.664 variants exhibit predominantly state-2-like conformations. FRET histograms for V1V4-tagged BG505 sgp140 SOSIP.664 with molecules after V3-negative selection (left), and for the stabilized BG505 sgp140 SOSIP.664 variant DS-SOSIP.Mut420 (right) (see Methods). Histograms represent mean ± s.e.m., determined from three independent populations of smFRET traces.
Extended Data Fig. 2 Binding of PGT151 stabilizes a state-2-like conformational state of HIV-1 Env.
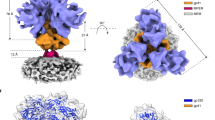
a, Structure of HIV-1JR-FL Env(ΔCT) in complex with PGT15119. Two PGT151 antigen-binding fragments are distant from the positions of the gp120 variable loops (V1 and V4) that carry the fluorophores. b, Population FRET histograms of unliganded HIV-1JR-FL Env(ΔCT), and HIV-1JR-FL Env(ΔCT) in the presence of 10 μg ml−1 sCD4D1D2–Igαtp. c–f, Addition of PGT151 at neutralizing concentrations (10 μg ml−1) shifts the conformational landscapes for enzymatically labelled HIV-1JR-FL (c, e) and HIV-1BG505 (d, f) from the unliganded preference towards state 1 (red solid lines) to a preference for state 2 (blue solid lines). g, The addition of PGT151 to BG505 sgp140 SOSIP.664 did not alter the dominance of the state-2-like conformation exhibited in the absence of PGT151. h, Schematic of use of amber-suppressor tRNAs to introduce unnatural amino acids that can be clicked with fluorophores (h, top; see Methods), and schematic comparison between the Q3 and A1 double tag used for enzymatically labelling and click-labelling of V1 and V4 of HIV-1JR-FL (h, middle) and HIV-1BG505 Env (h, bottom). To introduce the unnatural amino acid TCO*, Asn136 in the V1 loop of HIV-1JR-FL or Ser401 in the V4 loop of HIV-1BG505 was genetically altered to an amber (TAG) stop codon. i–l, Experiment as in c–f, characterizing the conformational landscape upon binding of PGT151 to click-labelled HIV-1JR-FL V1–Asn136TAG V4–A1 (i, k), and HIV-1BG505 V1–Q3 V4–Ser401TAG (j, l). Neutralization data (mean ± s.d.) are averaged from three independent experiments in triplicates (c, d, i, j). FRET population histograms represent mean ± s.e.m., determined from three independent populations of smFRET traces.
Extended Data Fig. 3 SOS and I559P effects on infectivity and conformational plasticity of sgp140 SOSIP.664.
a, SOS and/or I559P (IP) changes introduced into native HIV-1BG505 Q23 Env do not influence Env processing or virus incorporation. Env expression, processing and virus incorporation for HIV-1BG505 Q23 carrying SOS, I559P and SOS and I559P (SOS&IP) changes were tested by centrifugation of viruses from cell culture supernatants, followed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis in the presence of dithiothreitol, and western blotting using the antiserum to HIV-1 gp120 (NIH AIDS reagent no. 288) and HIV-1 p24 monoclonal antibody (NIH AIDS reagent no. 3537). Experiments were repeated twice. b, The structure-stabilizing modifications A501C and T605C (SOS) and I559P used in the design of BG505 sgp140 SOSIP.664 abort HIV-1 infectivity. Infectivity of HIV-1BG505 Q23 SOS and I559P was measured by a Gaussia Luciferase assay, and normalized to that of wild-type HIV-1BG505 Q23. c, FRET histogram of HIV-1JR-FL Env carrying SOS, confirming similar data for HIV-1BG505 Env that the SOS change is largely responsible for the state 2 stabilization of Env on virus. d, e, FRET histograms of HIV-1BG505 Env in the absence (unliganded, d) or the presence of sCD4D1D2–Igαtp (e). f, g, Experiments as in d, e for BG505 sgp140 SOSIP.664. h–k, FRET histograms of HIV-1BG505 Env and BG505 sgp140 SOSIP.664 in the presence of the entry inhibitors BMS-378806 (h, i) and BMS-626529 (j, k). l–m, Neutralization of HIV-1BG505 by sCD4D1D2–Igαtp (l), BMS-378806 (m) and BMS-626529 (n). Red arrows indicate concentrations used in smFRET experiments. Histograms correspond to those in the main figures: in d (Fig. 1e), e (Fig. 2b, top), f (Fig. 1f), g (Fig. 2c, top), j (Fig. 2b, bottom) and k (Fig. 2c, bottom). Infectivity and neutralization curves represent mean ± s.d. from three replicates in triplicates. FRET population histograms represent mean ± s.e.m., determined from three independent populations of smFRET traces.
Extended Data Fig. 4 Conformational remodelling of HIV-1BG505 and BG505 sgp140 SOSIP.664 by sCD4D1D2–Igαtp and BMS-626529.
a, b, Examples of fluorescence traces of BG505 sgp140 SOSIP.664 in the presence of 10 μg ml−1 sCD4D1D2–Igαtp (a), and 100 μM BMS-626529 (b). Arrows indicate single-step photobleaching events that define the background of our smFRET assay. c–e, Transition density plots of HIV-1BG505 in the absence (c) or presence (d) of sCD4D1D2–Igαtp, or in the presence of BMS-626529 (e). Transition density plots that indicate the relative frequency of state-to-state transitions were generated from individual traces (180 traces in Fig. 1e, 147 traces in Fig. 2b (top), and 116 traces in Fig. 2b (bottom)). n, number of total transitions observed. f–h, Transition density plots of BG505 sgp140 SOSIP.664 under the same experimental conditions as those shown in c–e.
Extended Data Fig. 5 Many bNAbs neutralize and exhibit preference for the state 1 conformation of HIV-1.
a–c, Neutralization of native HIV-1BG505 by bNAbs that recognize different Env epitopes: V3 glycan site-directed bNAbs 10-1074, PGT121 and PGT122 (a); CD4 binding site bNAbs 3BNC117, VRC01 and VRC03 (b); and V1V2 glycan bNAbs PG9, PG16 and PGT145 (c). Only bNAbs that potently neutralize HIV-1BG505 or HIV-1JR-FL and allowed smFRET imaging at an antibody concentration 5 times above the 95% inhibitory concentration were analysed further (Fig. 3b, d). Neutralization data (mean ± s.d.) were averaged from three independent experiments in triplicate. d, FRET histogram that shows that HIV-1BG505 Env remains in state 1 in the presence of PG9 (50 μg ml−1). FRET population histograms represent mean ± s.e.m., determined from three independent populations of smFRET traces.
Extended Data Fig. 6 Conformational preferences of non-neutralizing antibodies for HIV-1 Env on virus.
a, b, FRET histograms and overlaid landscapes of HIV-1NL4-3 in the presence of 100 μg ml−1 17b (a) and 100 μg ml−1 F105 (b) acquired after 0 min, 30 min and 60 min of incubation. c, d, FRET histograms and overlaid landscapes of HIV-1BG505 in the presence of 17b (c) and F105 (d), acquired after 0 min, 30 min and 60 min of incubation. Non-neutralizing antibodies have preference for the state 3 conformation of Env. Note that in contrast to the tier 1 HIV-1 isolate NL4-3, the tier 2 isolate BG505 does not respond to 17b. FRET population histograms represent mean ± s.e.m., determined from three independent groups of smFRET traces.
Extended Data Fig. 7 Antibodies isolated from cows immunized using BG505 sgp140 SOSIP.664 immunogens exhibit a preference for state 2.
a, FRET histogram of HIV-1BG505(T332N). b, Neutralization curves of HIV-1BG505 by NC-Cow1, NC-Cow8, NC-Cow9 and NC-Cow10 antibodies. Data are presented as mean ± s.d. determined from three independent experiments in triplicate. c–e, FRET histograms of native HIV-1BG505 in the presence of 10 μg ml−1 NC-Cow1 (c), NC-Cow8 (d) and NC-Cow10 (e). f, The FRET histogram of HIV-1BG505 that carries the T332N substitution in Env is overlaid with that of wild-type HIV-1BG505. The T332N substitution in HIV-1BG505 Env does not detectably change the conformation of the Env. g, h, Cow antibodies (NC-Cow1, NC-Cow8, NC-Cow9 and NC-Cow10) shift the conformational landscape of native Env on the virus from state 1 towards that of BG505 sgp140 SOSIP.664 (state 2). FRET population histograms (a, c–e) represent mean ± s.e.m., from three independent populations of smFRET traces. i, j, Ligand preferences for states 1 and 2 probed by antibody staining of cell-expressed HIV-1JR-FL Env(ΔCT). Increasing amounts of the first ligand were pre-bound to cells for 1 h. The cells were washed, incubated with the second dye-labelled probe for 30 min, and the binding was quantified by flow cytometry. The ratio of measured mean fluorescence intensity (MFI) was normalized to that seen in the absence of pre-bound ligand (Methods). Matched combinations (state 1 and state 1 or state 2 and state 2) and non-matched combinations (state 1 and state 2 or state 2 and state 1) at the highest concentration of pre-bound first ligand were compared, and statistical significance was evaluated using a paired Student’s two-sided t-test. *P < 0.05. Note that the strong interference between 3BNC117 and PGT151 is due to a steric clash between the two antibodies, and was included as a control.
Extended Data Fig. 8 Validating the behaviour of dyes used for smFRET.
a, The 50-ns molecular dynamics simulations of fluorophore tumbling on the BG505 sgp140 SOSIP.664 trimer (4ZMJ) shows that dyes in V1 and V4 are far from the viral membrane. Molecular dynamics simulation was performed to account for movements of loops, enzymatic labelling tags, linkers and dyes to describe the possible dye tumbling space within 50 ns. The sampled space was docked into the approximately 20 Å structure of the HIV-1 virus Env spike determined by cryo-electron tomography4. A 50-ns molecular dynamics simulation is not temporally comparable to the time resolution of single-molecule imaging at 40 ms, or the timescale of observed conformational changes of Env (milliseconds to seconds). b–d, Conformational properties of the HIV-1BG505 Env remain highly similar when the dyes are flipped. b, Reference FRET histograms of HIV-1BG505 that carries Cy3B in V1 and Cy5 in V4, in unliganded form (from Fig. 1e), in the presence of PGT151 (from Fig. 1h) or in the presence of sCD4D1D2–Igαtp (from Fig. 2b). c, FRET histograms of HIV-1BG505 Env that carries Cy5 in V1 and Cy3B in V4 (see Methods), in the absence and in the presence of 10 μg ml−1 PGT151 or 10 μg ml−1 sCD4D1D2–Igαtp, respectively. d, Overlaid conformational landscapes of HIV-1BG505 Env labelled as in c with flipped dyes (green), compared to HIV-1BG505 Env labelled as in b (red). FRET population histograms represent mean ± s.e.m., determined from three independent populations of smFRET traces.
Extended Data Fig. 9 Suppressed HIV-1JR-FL that carries amber positions in gp120 and gp41 enables smFRET imaging of Env from two distinct perspectives.
a, Schematic of tagged sites in HIV-1JR-FL Env that were used for enzymatic labelling and amber stop codon (TAG)-suppressed incorporation of unnatural amino acids for click labelling. HIV-1JR-FL V1–Q3 V4–A1 carries the Q3 peptide in the V1 loop and the A1 peptide in the V4 loop. HIV-1 JR-FL V1–Asn136TAG V4–A1 carries a TAG at position Asn136 in V1 and the A1 peptide in V4. HIV-1JR-FL V4–A1 α6–Arg542TAG carries the A1 tag in gp120 V4 and a TAG at Arg542 in the α6 helix of gp41. b, c, Neutralization of HIV-1JR-FL wild type, 100%-peptide-tagged V1–Q3 V4–A1, 100%-amber-suppressed V1–Asn136TAG V4–A1 and V4–A1 α6–Arg542TAG by sCD4D1D2–Igαtp (b), and eCD4-Ig(Q40A, mim2) (c). Neutralization curves (b, c) represent mean ± s.d. from three replicates in triplicates.
Supplementary information
Supplementary Table 1
This table shows the fluorescence lifetime of free and conjugated Cy3 and Cy5 used in smFRET imaging.
Rights and permissions
About this article
Cite this article
Lu, M., Ma, X., Castillo-Menendez, L.R. et al. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419 (2019). https://doi.org/10.1038/s41586-019-1101-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1101-y
This article is cited by
-
HIV-1 Env trimers asymmetrically engage CD4 receptors in membranes
Nature (2023)
-
Asymmetric conformations of cleaved HIV-1 envelope glycoprotein trimers in styrene-maleic acid lipid nanoparticles
Communications Biology (2023)
-
Intermediate conformations of CD4-bound HIV-1 Env heterotrimers
Nature (2023)
-
Structure-function analyses reveal key molecular determinants of HIV-1 CRF01_AE resistance to the entry inhibitor temsavir
Nature Communications (2023)
-
An HIV-1 broadly neutralizing antibody overcomes structural and dynamic variation through highly focused epitope targeting
npj Viruses (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.