Abstract
The non-canonical NF-κB signalling cascade is essential for lymphoid organogenesis, B cell maturation, osteoclast differentiation, and inflammation in mammals1,2; dysfunction of this system is associated with human diseases, including immunological disorders and cancer3,4,5,6. Although expression of NF-κB-inducing kinase (NIK, also known as MAP3K14) is the rate-limiting step in non-canonical NF-κB pathway activation2,7, the mechanisms by which transcriptional responses are regulated remain largely unknown. Here we show that the sine oculis homeobox (SIX) homologue family transcription factors SIX1 and SIX2 are integral components of the non-canonical NF-κB signalling cascade. The developmentally silenced SIX proteins are reactivated in differentiated macrophages by NIK-mediated suppression of the ubiquitin proteasome pathway. Consequently, SIX1 and SIX2 target a subset of inflammatory gene promoters and directly inhibit the trans-activation function of the transcription factors RELA and RELB in a negative feedback circuit. In support of a physiologically pivotal role for SIX proteins in host immunity, a human SIX1 transgene suppressed inflammation and promoted the recovery of mice from endotoxic shock. In addition, SIX1 and SIX2 protected RAS/P53-driven non-small-cell lung carcinomas from inflammatory cell death induced by SMAC-mimetic chemotherapeutic agents (small-molecule activators of the non-canonical NF-κB pathway). Our findings identify a NIK–SIX signalling axis that fine-tunes inflammatory gene expression programs under both physiological and pathological conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
All data generated during this study that support the findings are included in the manuscript or in its Source Data and Supplementary Information. All materials are available from authors upon reasonable request. The RNA-seq data associated with Fig. 4f and Extended Data Figs. 2b, 4g have been deposited in Gene Expression Omnibus (GEO) at NCBI (accession code: GSE126535)
References
Cildir, G., Low, K. C. & Tergaonkar, V. Noncanonical NF-κB signaling in health and disease. Trends Mol. Med. 22, 414–429 (2016).
Sun, S. C. The non-canonical NF-κB pathway in immunity and inflammation. Nat. Rev. Immunol. 17, 545–558 (2017).
Shinkura, R. et al. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-κb-inducing kinase. Nat. Genet. 22, 74–77 (1999).
Annunziata, C. M. et al. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007).
Keats, J. J. et al. Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007).
Rosebeck, S. et al. Cleavage of NIK by the API2–MALT1 fusion oncoprotein leads to noncanonical NF-κB activation. Science 331, 468–472 (2011).
Xiao, G., Harhaj, E. W. & Sun, S. C. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7, 401–409 (2001).
Coope, H. J. et al. CD40 regulates the processing of NF-κB2 p100 to p52. EMBO J. 21, 5375–5385 (2002).
Schoggins, J. W. et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 505, 691–695 (2014).
Silver, S. J. & Rebay, I. Signaling circuitries in development: insights from the retinal determination gene network. Development 132, 3–13 (2005).
Ford, H. L., Kabingu, E. N., Bump, E. A., Mutter, G. L. & Pardee, A. B. Abrogation of the G2 cell cycle checkpoint associated with overexpression of HSIX1: a possible mechanism of breast carcinogenesis. Proc. Natl Acad. Sci. USA 95, 12608–12613 (1998).
Vince, J. E. et al. TWEAK-FN14 signaling induces lysosomal degradation of a cIAP1–TRAF2 complex to sensitize tumor cells to TNFα. J. Cell Biol. 182, 171–184 (2008).
Varfolomeev, E. et al. IAP antagonists induce autoubiquitination of c-IAPs, NF-κB activation, and TNFα-dependent apoptosis. Cell 131, 669–681 (2007).
Vince, J. E. et al. IAP antagonists target cIAP1 to induce TNFα-dependent apoptosis. Cell 131, 682–693 (2007).
Kim, J. Y. et al. TNFα induced noncanonical NF-κB activation is attenuated by RIP1 through stabilization of TRAF2. J. Cell Sci. 124, 647–656 (2011).
Christensen, K. L., Brennan, J. D., Aldridge, C. S. & Ford, H. L. Cell cycle regulation of the human Six1 homeoprotein is mediated by APCCdh1. Oncogene 26, 3406–3414 (2007).
Li, X. et al. Eya protein phosphatase activity regulates Six1–Dach–Eya transcriptional effects in mammalian organogenesis. Nature 426, 247–254 (2003).
Patrick, A. N. et al. Structure-function analyses of the human SIX1–EYA2 complex reveal insights into metastasis and BOR syndrome. Nat. Struct. Mol. Biol. 20, 447–453 (2013).
Brown, K., Gerstberger, S., Carlson, L., Franzoso, G. & Siebenlist, U. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science 267, 1485–1488 (1995).
Wertz, I. E. et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430, 694–699 (2004).
Chew, J. et al. WIP1 phosphatase is a negative regulator of NF-κB signalling. Nat. Cell Biol. 11, 659–666 (2009).
Liu, B. et al. Negative regulation of NF-κB signaling by PIAS1. Mol. Cell. Biol. 25, 1113–1123 (2005).
Šošić, D., Richardson, J. A., Yu, K., Ornitz, D. M. & Olson, E. N. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-κB activity. Cell 112, 169–180 (2003).
Barboric, M., Nissen, R. M., Kanazawa, S., Jabrane-Ferrat, N. & Peterlin, B. M. NF-κB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8, 327–337 (2001).
McCoy, E. L. et al. Six1 expands the mouse mammary epithelial stem/progenitor cell pool and induces mammary tumors that undergo epithelial-mesenchymal transition. J. Clin. Invest. 119, 2663–2677 (2009).
Li, L. et al. A small molecule Smac mimic potentiates TRAIL- and TNFα-mediated cell death. Science 305, 1471–1474 (2004).
Petersen, S. L. et al. Autocrine TNFα signaling renders human cancer cells susceptible to Smac-mimetic-induced apoptosis. Cancer Cell 12, 445–456 (2007).
Cheung, H. H. et al. SMG1 and NIK regulate apoptosis induced by Smac mimetic compounds. Cell Death Dis. 2, e146 (2011).
Cheung, H. H., Mahoney, D. J., Lacasse, E. C. & Korneluk, R. G. Down-regulation of c-FLIP enhances death of cancer cells by Smac mimetic compound. Cancer Res. 69, 7729–7738 (2009).
Fulda, S. & Vucic, D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 11, 109–124 (2012).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Malinin, N. L., Boldin, M. P., Kovalenko, A. V. & Wallach, D. MAP3K-related kinase involved in NF-κB induction by TNF, CD95 and IL-1. Nature 385, 540–544 (1997).
Yin, L. et al. Defective lymphotoxin-β receptor-induced NF-κB transcriptional activity in NIK-deficient mice. Science 291, 2162–2165 (2001).
Zhang, X., Goncalves, R. & Mosser, D. M. The isolation and characterization of murine macrophages. Curr. Protoc. Immunol. 83, 14.1.1–14.1.14 (2008).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Tetsuka, T. et al. Inhibition of nuclear factor-κB-mediated transcription by association with the amino-terminal enhancer of split, a Groucho-related protein lacking WD40 repeats. J. Biol. Chem. 275, 4383–4390 (2000).
Perelman, S. S. et al. Cell-based screen identifies human interferon-stimulated regulators of Listeria monocytogenes infection. PLoS Pathog. 12, e1006102 (2016).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Aronesty, E. Comparison of sequencing utility programs. Open Bioinform. J. 7, 1–8 (2013).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Cormier, C. Y. et al. Protein Structure Initiative Material Repository: an open shared public resource of structural genomics plasmids for the biological community. Nucleic Acids Res. 38, D743–D749 (2010).
Dittmann, M. et al. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell 160, 631–643 (2015).
Bhattacharyya, S., Borthakur, A., Dudeja, P. K. & Tobacman, J. K. Lipopolysaccharide-induced activation of NF-κB non-canonical pathway requires BCL10 serine 138 and NIK phosphorylations. Exp. Cell Res. 316, 3317–3327 (2010).
Kumar, J. P. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell. Mol. Life Sci. 66, 565–583 (2009).
Acknowledgements
We thank H. Ford (University of Colorado) for the Tet-on HA-SIX1 transgenic embryos and for discussion in preparation of the manuscript; the UT Southwestern Medical Center transgenic core for reviving the frozen embryos; J. Minna (UTSW) for NSCLCs; and V. Tagliabracci, J. Mendell, D. Pan, I. D’Orso, N. Conrad, R. Brekken and members of the Alto Laboratory for discussions. This research was supported by grants from the National Institutes of Health (AI083359 to N.M.A., AI117922 to J.W.S., and 5K12 HD068369-09 to N.W.H.) and the Children’s Medical Center Foundation (N.W.H.), and grants to N.M.A. from the Welch Foundation (I-1731), The Burroughs Welcome Fund, and the Howard Hughes Medical Institute and Simons Foundation Faculty Scholars Program.
Reviewer information
Nature thanks Claus Scheidereit and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
Z.L. and N.M.A. conceived and designed the study. Unless otherwise specified, Z.L. performed all experiments. K.B.M. assisted with mouse experiments. N.W.H. performed West Nile Virus infection. S.S.P. performed RNA-seq to identify NIK-stimulated genes. M.K. and C.X. performed bioinformatics analysis of RNA-seq data. J.W.S. designed the genetic screening platform for bacteria and viruses and provided critical input into project directions. Z.L. and N.M.A. analysed data and wrote the manuscript with editorial input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 CD40-NIK signalling axis mediates antibacterial function.
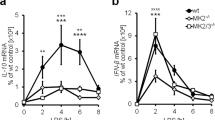
a, b, Experiments were performed to exclude the possibility that the observed CD40L-induced antibacterial function was specific to a particular cell type or protocol of cytokine induction. We reconstituted the CD40L signalling pathway in HEK293 cells. These cells do not express CD40, the endogenous receptor for CD40L (a, left). HEK293 cells are also unable to be stimulated by CD40L (a, right). However, overexpression of CD40 strongly induced NF-κB pathway activation (a, right). Expression of CD40 restricted both L. monocytogenes and S. flexneri infection (b) to levels similar to those observed in CD40L treated U-2 OS cells (compare with Fig. 1b). The NF-κB reporter activity assay (a, right) was performed by co-transfecting empty vector (EV), CD40L or CD40 with 5 × κB-LUC reporter gene into HEK293T cells. Luciferase activity was measured after 48 h and normalized to EV. Data are mean ± s.d. from six independent experiments. Experiment and quantification of b are presented as in Fig. 1b. Data are mean ± s.d. from six independent experiments. c, d, Confirmation of genetic knockout of the MAP3K14 (NIK) and MAP3K7 (TAK1) genes in STAT1−/− human fibroblasts. c, Schematic representation of In/Del base pairing and the sgRNA targets locus of exon 1 in NIK and TAK1. NIK−/− contains deletion of (−)7 bp, insertion of (+)G, and +CTCAC alleles (top). TAK1−/− contains +AT alleles, −2 bp, and −409 bp (bottom). d, Endogenous NIK and TAK1 expression in parental, NIK−/− and TAK1−/− cells. NIK is constitutively degraded by the cIAP–TRAF2/3 E3-ligase complex in quiescent cells2. Wild-type, NIK−/−, and TAK1−/− fibroblasts were treated with 2.5 μM BV6 for 14 h and then probed with indicated antibodies using western blotting. e, NIK is necessary for restricting L. monocytogenes infection. Fibroblasts with the indicated genetic background were treated with vehicle control (DMSO) or 2.5 μM BV6 for 14 h and then infected with LmGFP. Percentage bacterial infection was normalized to wild-type untreated control. Black asterisks denote the difference between wild-type and indicated cell lines; red denote the difference between DMSO and BV6 treatment. NIK−/− cells exhibited much higher L. monocytogenes infection than either wild-type or TAK−/− cells, consistent with the role of NIK in preventing infection after cellular stimulation. However, BV6 treatment—which suppressed L. monocytogenes infection of wild-type cells—had no effect on NIK−/− cells, further indicating that NIK activation is necessary for the antibacterial response. Data are mean ± s.d. from nine independent experiments. f, NIK kinase activity is required for its antibacterial function. Fluc, wild-type NIK or NIK-kinase dead mutant (NIKK429/430A referred to as NIKKD) lentivirus was transduced into fibroblasts or U-2 OS cells as indicated. Cells were then challenged with SfGFP. Quantification of bacterial infection is as in Fig. 1b. Data are mean ± s.d. from four independent experiments. g, NF-κB gene expression induced by NIK is kinase dependent. EV, NIK or NIKKD was co-transfected with 5 × κB-LUC into HEK293T cells. NF-κB activity was measured after 48 h and normalized to EV. Data are mean ± s.d. from four independent experiments. h, Expression of NIK, but not TAK1, inhibits infection with L. monocytogenes and S. flexneri. Wild-type U-2 OS cells were transduced with combinational FlucRFP/FlucBFP, NIKRFP/FlucBFP, or TAK1RFP/TAB1BFP lentivirus. Cells were then challenged with LmGFP or Sf GFP. Infection efficiency was quantified by flow cytometry, gating GFP+ cells in both RFP+ and BFP+ cell populations. The relative percentage of pathogen infection was normalized to Fluc control. Data are mean ± s.d. from eight independent experiments. i, NIK protein expression levels corresponding to experiments in Fig. 1c. Fluc- and NIK-transduced cells were lysed and probed with anti-NIK antibody. j, Previous studies have suggested that ectopic expression of NIK is cytotoxic in A549 cells44. To test whether ectopic expression of NIK is cytotoxic in fibroblasts, we transduced fibroblasts with indicated lentivirus and measured cell viability after 72 h by measuring ATP. Data are mean ± s.d. from six independent experiments. P values were measured using one-way ANOVA (GraphPad); ***P < 0.001, ****P < 0.0001; ns, no significant difference. The same statistics were used in later figures unless otherwise stated. Western blot data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Library screen for NIK-stimulated genes.
a, Schematic of NIK-stimulated gene library design, cloning, and multidimensional flow cytometry-based high-throughput screen. NIK-stimulated genes were identified by RNA-seq. The cDNAs of 237 NIK-stimulated genes were individually cloned into the lentiviral vector pTRIP upstream of IRES-tagRFP (see Methods). Fibroblasts were transduced with lentivirus in a one-gene to one-well format and were then infected with GFP-expressing pathogens, including Lm, Sf, EAV, WNV, SINV, or PIV3 in independent experiments. The effect of a single gene expression on infection was quantified by flow cytometry. b, Relative expression levels of NIK-stimulated genes identified by RNA-seq. Fluc or NIK lentivirus was transduced into fibroblasts. Total RNA was isolated after 72 h and gene expression was determined by RNA sequencing. Graph shows expression of genes that were significantly stimulated (red, 237 genes) or downregulated (green, 84 genes) by NIK expression compared to Fluc control. Fold change of more than 2 (log2 ≥ 1) or less than 0.5 (log2 ≤ −1) and FDR < 0.05 (see Methods). Bars were ranked numerically from low to high (see Supplementary Table 1). The expression levels of SIX1 and SIX2 are indicated. Data are representative of two independent experiments. c, Efficiency of lentiviral expression of NIK-stimulated genes used in the high-throughput bacterial and viral screen. NIK-stimulated genes were transduced into fibroblasts in a ‘one-gene to one-well’ format. Transduction efficiency measured as per cent RFP+ cells was determined by flow cytometry and was ranked numerically from low to high (see Source Data, values are the average of two technical replicates). Twelve out of 237 genes were poorly transduced (less than 20% RFP+) and were excluded from subsequent analyses. d, Dot plots of S. flexneri, L. monocytogenes, EAV, WNV, SINV, and PIV3 infectivity in the presence of expressed NIK-stimulated genes (in c). Data were normalized to the average of each screen, indicated as the black dotted line. We chose to confirm hits in Fig. 1d on the basis of two criteria: (1) the effect of gene expression on inhibiting or enhancing pathogen infection by less than or greater than 50%; and (2) an adjusted z-score less than −2 or greater than 2 (see Supplementary Table 2). NIK-stimulated genes that reproducibly and significantly inhibited (green) or enhanced infection (red) by these criteria are indicated. The genes shown in black font are hits that were not reproduced in the confirmatory experiments (Fig. 1d). Data are mean ± s.d. from two (S. flexneri, L. monocytogenes) or one (EAV, WNV, SINV, and PIV3) independent experiments.
Extended Data Fig. 3 NIK mediates reactivation of SIX proteins by inhibiting the ubiquitin–proteasome pathway.
a, Control experiment for Fig. 2a, c showing that NIK is expressed in wild-type BMDMs but not in BMDMs isolated from Nik−/− mice. NIK is constitutively degraded under quiescent condition. We used the MG132 proteasome inhibitor to stabilize endogenous NIK protein expression. To validate NIK protein expression, wild-type and Nik−/− primary BMDMs were mock-treated or treated with 30 μM MG132 for 12 h and NIK was detected by western blot. b, c, Long-term treatment of cells with TNF (b) or LPS45 (c) stabilized SIX1 expression through activation of NIK in mouse primary BMDMs. Wild-type and Nik−/− primary BMDMs were treated with 25 ng ml−1 TNF (b, quantification of SIX1 protein expression in TNF-treated cells, mean ± s.d. from three independent experiments as in Fig. 2c) or 100 ng ml−1 LPS (c) for 24 h. d, e, Reactivation of human SIX1 and SIX2 by long-term treatment with both canonical (TNF) and non-canonical (LTα1β2) NF-κB agonists requires NIK but not TAK1. Wild-type, NIK−/− or TAK1−/− fibroblasts were mock-treated or treated with 25 ng ml−1 TNF or 50 ng ml−1 LTα1β2 for 24 h. TAK1−/− cells were included as control to show that TNF and LTα1β2 could induce SIX1 and SIX2 accumulation in a TAK1-independent manner (e). f, g, Ectopic expression of NIK induces expression of recombinant SIX1 and SIX2 driven by the strong CMV promoter in HEK293 cells. Plasmids encoding CMV-driven GFP–SIX1 or GFP–SIX2 were co-transfected into HEK293T cells with empty vector or Flag–NIK. Western blot (f) and fluorescence microscopy (g) assays were performed to detect expression of GFP–SIX1 and GFP–SIX2 48 h after transfection. We estimate that SIX1 and SIX2 protein are expressed in 5–10% of untreated cells and 60–70% of cells when co-transfected with NIK. Microscopy images were taken using a 10× objective, scale bars represent 150 μm (g). h, i, Activation of NIK by BV6 (h) or by inhibition of the proteasome with MG132 (i) stabilizes CMV–Flag–SIX2 expression in HEK293T cells. Flag–SIX2 was transfected into HEK293T cells for 24 h and cells were mock-treated or treated with 5 μM BV6 for 24 h or 30 μM MG132 for 12 h. j, Inhibition of the 26S proteasome with 30 μM MG132 induces endogenous SIX1 protein expression in primary BMDMs. k, Inhibition of the 26S proteasome promotes SIX1 and SIX2 expression in human fibroblasts, including NIK−/− fibroblasts (as in j). l, Kinetics of cIAP1 degradation and NIK, SIX1 and SIX2 accumulation in human fibroblasts treated with 5 μM BV6 for the indicated time. m, NIK suppresses SIX2 ubiquitination. HEK293T cells were co-transfected with HA–ubiquitin, Flag–SIX2 and GFP–NIK as indicated and cells were incubated for 48 h. SIX2 was immunoprecipitated with an anti-Flag antibody. The ubiquitination status of the protein was determined by anti-HA western blot. n, Reactivation mechanism of SIX proteins in response to non-canonical NF-κB activation (see main text). All data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 4 SIX proteins oppose NIK-mediated antibacterial function by inhibiting NIK-stimulated gene expression.
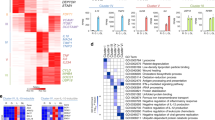
a, Previous studies have shown that SIX proteins assemble gene co-activator complexes through interaction with EYA family members17 and that SIX1 residues C16 and V17 are required for this interaction18. These residues are conserved in SIX2. We used mutant SIX2(C16RV17E) to show that SIX2 enhances L. monocytogenes infection independent of EYA interaction. Fluc, wild-type SIX2, or SIX2C16RV17E lentivirus was transduced into fibroblast cells. Cells were then challenged with LmGFP. The percentage infection was normalized to Fluc control. Data are mean ± s.d. from four independent experiments. b, Expression of SIX2 suppresses the antimicrobial function of NIK and CD40. Fibroblasts were lentivirally transduced with a combination of cDNAs (FlucRFP/FlucBFP, NIKRFP/FlucBFP, NIKRFP/SIX2BFP, CD40RFP/FlucBFP, or CD40RFP/SIX2BFP). After 72 h, cells were infected with LmGFP. RFP-, BFP- and GFP-expressing cells were gated by flow cytometry. Infection was quantified as in Extended Data Fig. 1h. Data are mean ± s.d. from six independent experiments. ***P < 0.001, ****P < 0.0001; ns, no significant difference. c, d, Characterization of SIX1−/−SIX2−/− fibroblasts generated by CRISPR–Cas9. c, Schematic representation of In/Del base pairing and the sgRNA that targets locus of exon 1 in the SIX1 and SIX2 genes in fibroblasts. SIX1−/−SIX2−/− contains a single T insertion in both alleles of the SIX1 and SIX2 genes. d, Western blot shows endogenous SIX1 and SIX2 expression in parental and SIX1−/−SIX2−/− fibroblasts. Data are representative of three independent experiments. e, The antibacterial activity of NIK is enhanced in SIX1−/−SIX2−/− fibroblasts. Wild-type and SIX1−/−SIX2−/− fibroblasts were transduced with Fluc or NIK lentivirus. After 72 h, cells were challenged with LmGFP. Black asterisks denote the difference between WT (Fluc) and WT (NIK) or SIX1−/−SIX2−/− (Fluc or NIK). Red asterisks denote the difference between WT (NIK) and SIX1−/−SIX2−/− (NIK). Relative infectivity was normalized to WT (Fluc) control. These data indicate that SIX proteins oppose the function of NIK, potentially through suppression of non-canonical NF-κB gene expression (see f). Data are presented as mean ± s.d. from six independent experiments. ****P < 0.0001; ns: no significant difference. f, Relationship between NIK expression, SIX protein accumulation, and antimicrobial immunity. g, RNA-seq experiments used to identify NIK-stimulated genes that are suppressed by SIX2. The indicated combination of lentiviruses (group I: FlucRFP and FlucBFP; group II: NIKRFP and FlucBFP; group III: FlucRFP and SIX2BFP; group IV: NIKRFP and SIX2BFP)were transduced into fibroblasts. Total RNA was extracted for deep sequencing 72 h later (Supplementary Table 3). h, Group comparisons from data generated in g. In brief, NIK-stimulated genes that are suppressed by SIX2 were determined by comparing Group IV with Group II (log2 ≤ −1) and then adjusted to NIK-stimulated genes from comparison of Group II with Group I (fold change > 4), FDR < 0.01 (see Methods). i, Representative raw data from RNA-seq experiments in g, h presented as fragments per kilobase of transcript per million mapped reads (FPKM) (bars show mean from two independent experiments indicated as circles). j, Validation of RNA-seq data. Experiments were performed as in g. Gene transcription level was determined by qRT–PCR and relative gene expression was normalized to Fluc control. Data are mean ± s.d. of three or two technical replicates and representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 5 SIX family proteins inhibit RELA- and RELB-mediated NF-κB activation.
a, b, Characterization of RELA−/− and RELB−/− human fibroblasts. Schematic representation of In/Del base pairing and the sgRNA targeting locus of exon 3 in RELA and exon 1 in RELB. a, Top, RELA−/− cells contain +C, −371 bp, and a 109-bp fragment that was replaced with 122 bp containing GCGCTA with reverse complementarity (orange). a, Bottom, RELB−/− cells contain the indicated deletions. b, Western blot shows endogenous RELA and RELB expression in parental and RELA−/− or RELB−/− fibroblasts and response to stimulation by 24-h application of TWEAK. c, Contributions of the canonical and non-canonical NF-κB subunits RELA and RELB, respectively, to gene expression. Fluc or NIK was transduced into wild-type, RELA−/−, and RELB−/− fibroblasts to stimulate the non-canonical NF-κB signalling pathway. mRNA expression of the indicated genes was evaluated by qRT–PCR. We conclude that IL1B is specifically stimulated by RELA because it is expressed in RELB−/− cells but not in RELA−/− cells. Similar logic was used to evaluate the seven additional genes shown. Experiments were performed as in Fig. 2g. Bars are means of two technical replicates (shown as circles) and representative of two independent experiments. d, The human SIX family consists of six unique isoforms. To determine which of these genes suppress NF-κB-mediated gene expression, empty vector (EV), SIX1, SIX3, SIX4, SIX5, or SIX6 cDNA was co-transfected with 5 × κB-LUC into HEK293T cells. After 24 h, cells were mock-treated or treated with 25 ng ml−1 TNF for 24 h. Luciferase activity was measured and normalized to EV untreated control. SIX1, SIX3, and SIX6 (SIX2 was not evaluated) inhibited both basal and inducible activity of NF-κB. Data are mean ± s.d. from seven independent experiments. e, SIX2 inhibits LTα1β2- and TNF-induced NF-κB activation. EV or Flag–SIX2 was co-transfected with 5 × κB-LUC into fibroblasts. After 24 h, cells were mock-treated or treated with 25 ng ml−1 TNF or 50 ng ml−1 LTα1β2 for 24 h. Data were analysed as in d. Data are mean ± s.d. from six independent experiments. f, The inhibitory potency of SIX2 is equivalent to those of A20 and IκBαSR. The Flag-tagged genes indicated were co-transfected with 5 × κB-LUC into HEK293T cells. After 24 h, cells were mock-treated or treated with 25 ng ml−1 TNF for 24 h. Data were analysed as in d. Data are mean ± s.d. from nine independent experiments. Anti-Flag western blot was performed to determine expression levels; IκBα regulation was included as a pathway activation control upon cellular stimulation with TNF. g, SIX2 inhibits the transcriptional activity of RELA and RELB. To evaluate the transcriptional activity of each NF-κB isoform independently, we transfected RELA cDNA into RELB−/− cells and RELB cDNA into RELA−/− cells along with SIX2 and 5 × κB-LUC. Transfection of either RELA or RELB potently induced NF-κB transcription of the indicated cells, and this transcription was suppressed by SIX2. Luminescence units were measured 48 h after transfection. Data are mean ± s.d. from twelve (left) and nine (right) independent experiments. All western blot data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 6 SIX family proteins inhibit NF-κB activation by occupying the target gene promoters.
a, Wild-type and SIX1−/−SIX2−/− H1792 cells were treated with mock or 50 ng ml−1 TWEAK for 24 h. Cell lysates from cytoplasmic or nuclear fractions were analysed by western blot. Neither SIX1 nor SIX2 blocked RELA phosphorylation or p100/52 processing, or restricted NF-κB translocation to the nucleus. b, Top, SIX proteins are composed of a SIX domain (SD), homeobox domain (HD), and coiled-coil (CD) region. Full-length SIX1 and SIX2 have 80% identical amino acids over the protein-coding sequence. The SD and HD domains (residues 1–183) are 96% identical46. The indicated Flag-tagged SIX2 fragments were transfected alone into U-2 OS cells and processed for microscopy (middle, scale bars represent 20 μm) or transfected with 5 × κB-LUC into HEK293T cells and processed for κB reporter activity (bottom). The highly conserved SD–HD domain was the minimal fragment that inhibited κB reporter activity. This fragment strictly localized to the nucleus, indicating that SIX proteins inhibit nuclear activity of NF-κB. Graph data analysed as in Extended Data Fig. 5d. Data are mean ± s.d. from nine independent experiments. c, SIX2 inhibits gene activation from the IL8 promoter. pIL8-LUC plasmid composed of the 1.5-kb promoter region of IL8 cloned upstream of LUC was co-transfected with indicated plasmids into HEK293T cells. After 24 h, cells were mock-treated or treated with 25 ng ml−1 TNF for 24 h. Luciferase activity was then measured and analysed as in Extended Data Fig. 5d. Data are mean ± s.d. from nine independent experiments. These data suggest that SIX proteins inhibit NF-κB activation at gene promoters. d, ChIP experiment providing additional evidence that SIX proteins bind to inflammatory gene promoters (Fig. 3a). Chromatin was prepared from stable GFP–SIX1 cell lines (HCT116 cells), mock-treated or treated with 25 ng ml−1 TNF for 2 h. Anti-SIX1 antibodies (or anti-IgG control) were used to immunoprecipitate SIX1 from nuclear extracts. Co-eluted DNA was amplified using primer sets as in Fig. 3a. Relative promoter occupancy was normalized to each experimental IgG control. Bars are means of two technical replicates (shown as circles) and data are representative of three independent experiments. e, Control experiments corresponding to Fig. 3a and Extended Data Fig. 6f showing GFP–SIX1 expression. Wild-type and stable GFP–SIX1 fibroblasts were stimulated with 50 ng ml−1 TWEAK for 24 h. GFP–SIX1 expression was measured by western blot. f, SIX1 expression does not affect recruitment of RELA to the IL8 promoter. Chromatin was prepared from wild-type or GFP–SIX1 stable fibroblasts and immunopreciptated with anti-RELA. Bound DNA was amplified and quantified by qPCR. Results were adjusted to ‘input DNA’ that was saved before immunopreciptation. Relative enrichment was then normalized to each group’s untreated control. Data are mean ± s.d. from three independent experiments. g, Control experiments corresponding to Fig. 3c showing that SIX2 inhibits 5 × κB-LUC activity induced by GAL4–RELA and GAL4–RELB. GAL4 –RELA or GAL4–RELB construct was co-transfected with indicated plasmids into HEK293T cells. Forty-eight hours after transfection, the luminescence units were measured. Data are mean ± s.d. from nine (left) and twelve (right) independent experiments. h, Model showing NIK-mediated reactivation of SIX protein function in a negative feedback loop to control inflammatory gene expression by targeting gene promoters and inhibiting the trans-activation function of NF-κB. In quiescent cells (top), NIK and SIX are constitutively ubiquitinated and degraded by the proteasome. Non-canonical NF-κB agonists (for example, TWEAK, LTα1β2, or BV6) promote degradation of cIAPs, loss of NIK ubiquitination and subsequent NIK protein accumulation (middle). Stabilized NIK activates non-canonical NF-κB-mediated inflammatory gene expression (middle). Under conditions of long-term cytokine exposure, NIK-mediated suppression of a currently unknown E3-ubiquitin ligase results in SIX protein accumulation (bottom). Consequently, SIX proteins suppress inflammatory gene expression by targeting gene promoters and directly inhibiting trans-activation by NF-κB in a negative feedback loop (bottom). All western blots and microscopy data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 7 Doxycycline-induced HA–SIX1 expression in mice.
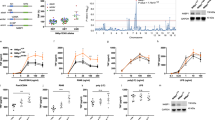
a, Doxycycline-induced HA–SIX1 bitransgenic mouse model system. CAG–rtTA3 mice were intercrossed with Tet-on driven HA–SIX1 mice to obtain rtTA3+/−SIX1+ mice. In principle, doxycycline-bound rtTA3 targets the Tet-on operator and drives broad HA–SIX1 expression across multiple tissues. Primer sets used to genotype CAG–rtTA3 on chromosome 8 and Tet-on HA–SIX1 are shown. Electrophoresis gels show genotyping of a representative rtTA3+/−SIX1+ mouse. b, Anti-SIX1 western blot of whole cell lysates from BMDMs isolated from rtTA3+/−SIX1+ mice. HA–SIX1 is not expressed in the absence of doxycycline under quiescent condition (left). When TNF was administered to these cells, endogenous SIX1 was stimulated (right). c, Whole cell lysates from doxycycline-treated BMDMs isolated from rtTA3+/− or rtTA3+/−SIX1+ mice. BMDMs were stimulated with TNF as indicated and probed with anti-SIX1 antibody by western blot. Doxycycline induced HA–SIX1 expression (lane 1), and this induction was potentiated by TNF (lane 2). HA–SIX1 ran as a doublet, which potentially represents an unmodified and a mono-ubiquitinated form of SIX1. Neither endogenous SIX1 nor HA–SIX1 was detected in BMDMs isolated from Dox-treated rtTA3+/− mice (lane 3). In control experiments, TNF induced endogenous SIX1 in BMDMs isolated from Dox-treated rtTA3+/− mice (lane 4). d, rtTA3+/−SIX1+ and rtTA3+/− mice were given 2 mg ml−1 doxycycline in drinking water for 10 days. Cell lysates from liver and spleen were used to probe HA–SIX1 expression by anti-SIX1 western blot. All western blot data are representative of three independent experiments. e, Peritoneal macrophages were isolated from rtTA3+/−SIX1+ and rtTA3+/− littermate control mice. Adherent macrophages were incubated with 2 μg ml−1 doxycycline for 24 h and then mock-treated or treated with 100 ng ml−1 LPS for 4 h. Total RNA was isolated for qRT–PCR. Relative gene expression was normalized to rtTA3+/− untreated control. Bars show mean from two technical replicates (shown as circles). Data are representative of three independent experiments. f, Experimental procedures corresponding to Fig. 4a–c (see Methods). For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 8 NIK−/− and SIX1−/−SIX2−/− fibroblasts are sensitized to BV6–TNF-induced cell death.
We validated the observation that SIX1−/−SIX2−/− sensitized NSCLC cells to cell death induced by BV6 and TNF in SV40 immortalized STAT1−/− fibroblasts. a, b, Fibroblasts of the indicated genotype were treated with BV6 (a) or TNF (b). Cell viability was determined by measuring ATP after 24 h. Cell survival rate was normalized to each genotype’s untreated control. Neither BV6 nor TNF (10 ng ml−1) alone induced fibroblast cell death. c, NIK−/− and SIX1−/−SIX2−/− fibroblasts are sensitized to BV6–TNF-induced cell death. Left, experiments and analysis as in a. Right, representative images showing the cell death phenotype induced by BV6–TNF in fibroblasts of the indicated genotypes. Scale bars represent 50 μm. d, Time course of combined BV6 (2.5 μM) and TNF (25 ng ml−1) treatment. NIK−/− and SIX1−/−SIX2−/− fibroblasts exhibited increased cleavage of poly ADP-ribose polymerase (PARP) and caspase-3 in BV6–TNF-treated fibroblasts. BV6–TNF induced NIK-dependent expression of both SIX1 and SIX2, suggesting that this cascade may be responsible for resistance to this treatment. e, f, We introduced a silent mutation in the gRNA recognition sequence of SIX2 cDNA that cannot be targeted by CRISPR–Cas9 (SIX2R, e, top). Expression of SIX2R in SIX1−/−SIX2−/− fibroblasts rescued the cell death phenotype (e) and suppressed both PARP and caspase-3 cleavage (f) induced by BV6–TNF. Wild-type and SIX1−/−SIX2−/− fibroblasts were transduced with Fluc or SIX2R lentivirus. After 72 h, cells were mock-treated or treated with 0.2 μM BV6 plus 10 ng ml−1 TNF for 24 h. Cell viability was determined by measuring ATP. Cell survival was normalized to each untreated control (e, bottom). For western blot, cells were treated with 2.5 μM BV6 plus 25 ng ml−1 TNF for 6 h (f). All quantified data are mean ± s.d. from nine independent experiments. **P < 0.01, ****P < 0.0001. Western blot data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 9 NIK−/− and SIX1−/−SIX2−/− sensitize U-2 OS cells to BV6–TNF-induced caspase-8-dependent cell death.
a–d, Knockout of NIK or SIX1 and SIX2 in U-2 OS cells using the CRISPR–Cas9 system. Western blots show endogenous NIK, SIX1, or SIX2 expression in parental, NIK−/− and SIX1−/−SIX2−/− U-2 OS cells (a, c). We used MG132 to stabilize endogenous NIK in wild-type and NIK−/− U-2 OS cells. b, Schematic representation of In/Del base pairing and the sgRNA targeting locus of exon 1 in NIK in U-2 OS cells. NIK−/− contains +G, −5 bp, and −18 bp (disrupted alternative splicing site) alleles. d, Schematic representation of In/Del base pairing and the sgRNA targeting locus of exon 1 in SIX1 and SIX2 in U-2 OS cells. SIX1−/−SIX2−/− contains +T of SIX1 and SIX2. e, SIX1−/−SIX2−/− and NIK−/− U-2 OS cells are sensitive to BV6–TNF-induced apoptosis. Wild-type, NIK−/−, or SIX1−/−SIX2−/− U-2 OS cells were treated with 25 ng ml−1 TNF alone or in the presence of indicated concentrations of BV6 for 48 h. Cell viability was measured by ATP. Cell survival was normalized to the absence of BV6 (control). f, g, Expression of SIX2R (see Extended Data Fig. 8e) or SIX2 cDNA protected SIX1−/−SIX2−/− or NIK−/− cells, respectively, from BV6–TNF-induced apoptosis. Wild-type, NIK−/−, or SIX1−/−SIX2−/− U-2 OS cells were transduced with Flu, SIX2 or SIX2R lentivirus as indicated and treated with 2.5 μM BV6 plus 25 ng ml−1 TNF for 24 h. h, j, Cell death was mediated by the extrinsic apoptotic pathway, as both the pan-caspase inhibitor z-VAD and specific caspase-8 inhibitor z-IETD blocked BV6–TNF-induced cell death in SIX1−/−SIX2−/− fibroblasts (h) and U-2 OS cells (j). Wild-type and SIX1−/−SIX2−/− fibroblasts were treated with 1 μM BV6 plus 10 ng ml−1 TNF alone or in the presence of 20 μM z-VAD or z-IETD for 24 h. U-2 OS cells were treated with 2.5 μM BV6 plus 25 ng ml−1 TNF alone or in the presence of 20 μM z-VAD for 48 h. Cell viability was measured by ATP. Cell survival rate was normalized to each untreated control. i, Western blot showing the status of PARP and caspase-3 cleavage induced by BV6–TNF treatment in the absence of or the presence of caspase inhibitors in wild-type and SIX1−/−SIX2−/− fibroblasts. Cells were mock-treated or treated with 2.5 μM BV6 plus 25 ng ml−1 TNF alone or in the presence of 30 μM z-VAD or 40 μM z-IETD for 6 h. All quantified data are mean ± s.d. from nine (e, f, h, j) or six (g) independent experiments. *P < 0.05, ****P < 0.0001. Western blot data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 10 Validation and pathway analysis of RNA-seq in wild-type and SIX1−/−SIX2−/− H1792 NSCLC cells.
a, To validate cluster I genes from RNA-seq data shown in Fig. 4f, wild-type and SIX1−/−SIX2−/− H1792 cells were mock-treated or treated with 5 μM BV6 plus 25 ng ml−1 TNF for 24 h. Total RNA was isolated for qRT–PCR. Gene expression was normalized to wild-type untreated control. Data are mean ± s.d. of three technical replicates and are representative of three independent experiments. b, Ingenuity pathway analysis (IPA) of cluster I genes was performed (see Methods). Pathway enrichment bar plots are shown. Data are from two independent experiments. Significance values for the canonical pathways were calculated by Fisher’s exact test (right-tailed). c, Inflammatory gene transcription is highly induced in SIX1−/−SIX2−/− H1792 cells (compared to wild-type controls) upon specific stimulation of the non-canonical NF-κB signalling pathway. Wild-type and SIX1−/−SIX2−/− H1792 cells were mock-treated or treated with 50 ng ml−1 TWEAK for 3 h. Total RNA was extracted for qRT–PCR. Gene expression level was normalized to wild-type untreated control. Data are mean ± s.d. of three technical replicates and representative of two independent experiments.
Supplementary information
Supplementary Figures
This file contains uncropped data scans.
Supplementary Table
This file contains Supplementary Table S1: NIK-stimulated and downregulated genes.
Supplementary Table
This file contains Supplementary Table S2: NIK-stimulated genes screen datasets.
Supplementary Table
This file contains Supplementary Table S3: SIX2 downregulated NIK-stimulated genes (corresponds to Extended Data Fig. 4g, h).
Supplementary Table
This file contains Supplementary Table S4: Cluster I (up) (corresponds to Fig. 4f left panel).
Supplementary Table
This file contains Supplementary Table S5: Cluster II (no change) (corresponds to Fig. 4f middle panel).
Supplementary Table
This file contains Supplementary Table S6: Cluster III (down) (corresponds to Fig. 4f right panel).
Source data
Rights and permissions
About this article
Cite this article
Liu, Z., Mar, K.B., Hanners, N.W. et al. A NIK–SIX signalling axis controls inflammation by targeted silencing of non-canonical NF-κB. Nature 568, 249–253 (2019). https://doi.org/10.1038/s41586-019-1041-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1041-6
This article is cited by
-
NF-κB in biology and targeted therapy: new insights and translational implications
Signal Transduction and Targeted Therapy (2024)
-
Hierarchical graph neural network with subgraph perturbations for key gene cluster discovery in cancer staging
Complex & Intelligent Systems (2024)
-
Differential usage of DNA modifications in neurons, astrocytes, and microglia
Epigenetics & Chromatin (2023)
-
Retinal determination gene networks: from biological functions to therapeutic strategies
Biomarker Research (2023)
-
Myeloid-derived growth factor alleviates non-alcoholic fatty liver disease alleviates in a manner involving IKKβ/NF-κB signaling
Cell Death & Disease (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.