Abstract
The establishment of cell types during development requires precise interactions between genes and distal regulatory sequences. We have a limited understanding of how these interactions look in three dimensions, vary across cell types in complex tissue, and relate to transcription. Here we describe optical reconstruction of chromatin architecture (ORCA), a method that can trace the DNA path in single cells with nanoscale accuracy and genomic resolution reaching two kilobases. We used ORCA to study a Hox gene cluster in cryosectioned Drosophila embryos and labelled around 30 RNA species in parallel. We identified cell-type-specific physical borders between active and Polycomb-repressed DNA, and unexpected Polycomb-independent borders. Deletion of Polycomb-independent borders led to ectopic enhancer–promoter contacts, aberrant gene expression, and developmental defects. Together, these results illustrate an approach for high-resolution, single-cell DNA domain analysis in vivo, identify domain structures that change with cell identity, and show that border elements contribute to the formation of physical domains in Drosophila.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data tables containing the 3D positions of the barcodes localized by ORCA are available from the online repository: https://github.com/BoettigerLab/BXC-ORCA-data. An annotated map of the domains studied in this work is available as a BED file in the Supplementary Data and can also be viewed through the UCSC genome browser at https://tinyurl.com/y7wf8t6x. Additional microscopy data are not hosted online owing to their size (12.33 Tb), but are available upon reasonable request.
References
Long, H. K., Prescott, S. L. & Wysocka, J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187 (2016).
Spielmann, M., Lupiáñez, D. G. & Mundlos, S. Structural variation in the 3D genome. Nat. Rev. Genet. 19, 453–467 (2018).
Furlong, E. E. M. & Levine, M. Developmental enhancers and chromosome topology. Science 361, 1341–1345 (2018).
Schwartz, Y. B. & Cavalli, G. Three-dimensional genome organization and function in Drosophila. Genetics 205, 5–24 (2017).
Rowley, M. J. & Corces, V. G. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 19, 789–800 (2018).
Chambeyron, S. & Bickmore, W. A. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 18, 1119–1130 (2004).
Eskeland, R. et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol. Cell 38, 452–464 (2010).
Bantignies, F. et al. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell 144, 214–226 (2011).
Beliveau, B. J. et al. in Super-Resolution Microscopy: Methods and Protocols (ed. Erfle, H.) 231–252 (Springer, New York, 2017).
Williamson, I., Lettice, L. A., Hill, R. E. & Bickmore, W. A. Shh and ZRS enhancer colocalisation is specific to the zone of polarising activity. Development 143, 2994–3001 (2016).
Cheutin, T. & Cavalli, G. Loss of PRC1 induces higher-order opening of Hox loci independently of transcription during Drosophila embryogenesis. Nat. Commun. 9, 3898 (2018).
Cattoni, D. I. et al. Single-cell absolute contact probability detection reveals chromosomes are organized by multiple low-frequency yet specific interactions. Nat. Commun. 8, 1753 (2017).
Boettiger, A. N. et al. Super-resolution imaging reveals distinct chromatin folding for different epigenetic states. Nature 529, 418–422 (2016).
Kundu, S. et al. Polycomb repressive complex 1 generates discrete compacted domains that change during differentiation. Mol. Cell 65, 432–446 (2017).
Fabre, P. J. et al. Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states. Proc. Natl Acad. Sci. USA 112, 13964–13969 (2015).
Szabo, Q. et al. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci. Adv. 4, eaar8082 (2018).
Bintu, B. et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362, eaau1783 (2018).
Nir, G. et al. Walking along chromosomes with super-resolution imaging, contact maps, and integrative modeling. PLoS Genet. 14, e1007872 (2018).
Wang, S. et al. Spatial organization of chromatin domains and compartments in single chromosomes. Science 353, 598–602 (2016).
Cardozo Gizzi, A. M. et al. Microscopy-based chromosome conformation capture enables simultaneous visualization of genome organization and transcription in intact organisms. Mol. Cell https://doi.org/10.1016/j.molcel.2019.01.011 (2019).
Chen, K. H., Boettiger, A. N., Moffitt, J. R., Wang, S. & Zhuang, X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015).
Beliveau, B. J. et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc. Natl Acad. Sci. USA 109, 21301–21306 (2012).
Schuettengruber, B. et al. Cooperativity, specificity, and evolutionary stability of Polycomb targeting in Drosophila. Cell Rep. 9, 219–233 (2014).
Bonev, B. et al. Multiscale 3D genome rewiring during mouse neural development. Cell 171, 557–572 (2017).
Kyrchanova, O. et al. The boundary paradox in the Bithorax complex. Mech. Dev. 138, 122–132 (2015).
Maeda, R. K. & Karch, F. The bithorax complex of Drosophila. An exceptional Hox cluster. Curr. Top. Dev. Biol. 88, 1–33 (2009).
Maeda, R. K. & Karch, F. The open for business model of the bithorax complex in Drosophila. Chromosoma 124, 293–307 (2015).
Rivera, J., Keränen, S. V. E., Gallo, S. M. & Halfon, M. S. REDfly: the transcriptional regulatory element database for Drosophila. Nucleic Acids Res. 47 (D1), D828–D834 (2019).
Symmons, O. et al. Functional and topological characteristics of mammalian regulatory domains. Genome Res. 24, 390–400 (2014).
Symmons, O. et al. The Shh topological domain facilitates the action of remote enhancers by reducing the effects of genomic distances. Dev. Cell 39, 529–543 (2016).
Levine, M., Cattoglio, C. & Tjian, R. Looping back to leap forward: transcription enters a new era. Cell 157, 13–25 (2014).
Chen, H. et al. Dynamic interplay between enhancer-promoter topology and gene activity. Nat. Genet. 50, 1296–1303 (2018).
Bender, W. & Hudson, A. P element homing to the Drosophila bithorax complex. Development 127, 3981–3992 (2000).
Bowman, S. K. et al. H3K27 modifications define segmental regulatory domains in the Drosophila bithorax complex. eLife 3, e02833 (2014).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Lewis, E. B. A gene complex controlling segmentation in Drosophila. Nature 276, 565–570 (1978).
Rowley, M. J. et al. Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852 (2017).
Bender, W. & Lucas, M. The border between the ultrabithorax and abdominal-A regulatory domains in the Drosophila bithorax complex. Genetics 193, 1135–1147 (2013).
Gyurkovics, H., Gausz, J., Kummer, J. & Karch, F. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 9, 2579–2585 (1990).
Gambetta, M. C. & Furlong, E. E. M. The insulator protein CTCF is required for correct Hox gene expression, but not for embryonic development in Drosophila. Genetics 210, 129–136 (2018).
The modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Ghavi-Helm, Y. et al. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512, 96–100 (2014).
Rao, S. S. P. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Kragesteen, B. K. et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat. Genet. 50, 1463–1473 (2018).
Andrey, G. et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167 (2013).
Phanstiel, D. H. et al. Static and dynamic DNA loops form AP-1-bound activation hubs during macrophage development. Mol. Cell 67, 1037–1048 (2017).
Narendra, V. et al. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science 347, 1017–1021 (2015).
Narendra, V., Bulajić, M., Dekker, J., Mazzoni, E. O. & Reinberg, D. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 30, 2657–2662 (2016).
Van Bortle, K. et al. Insulator function and topological domain border strength scale with architectural protein occupancy. Genome Biol. 15, R82 (2014).
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods 5, 159–161 (2008).
Moffitt, J. R. et al. High-throughput single-cell gene-expression profiling with multiplexed error-robust fluorescence in situ hybridization. Proc. Natl Acad. Sci. USA 113, 11046–11051 (2016).
Gu, B. et al. Transcription-coupled changes in nuclear mobility of mammalian cis-regulatory elements. Science 359, 1050–1055 (2018).
Moffitt, J. R. et al. High-performance multiplexed fluorescence in situ hybridization in culture and tissue with matrix imprinting and clearing. Proc. Natl Acad. Sci. USA 113, 14456–14461 (2016).
Babcock, H., Sigal, Y. M. & Zhuang, X. A high-density 3D localization algorithm for stochastic optical reconstruction microscopy. Opt. Nanoscopy 1, 6 (2012).
Perry, M. W., Boettiger, A. N., Bothma, J. P. & Levine, M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr. Biol. 20, 1562–1567 (2010).
Briggs, J. A. et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018).
Van Der Maaten, L. J. P. & Hinton, G. E. Visualizing high-dimensional data using t-sne. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Ester, M., Kriegel, H.-P., Sander, J. & Xu, X. A density-based algorithm for discovering clusters. KDD-96 Proc. 226–231 (1996).
Heris, S. DBSCAN Clustering Algorithm https://www.mathworks.com/matlabcentral/fileexchange/52905-dbscan-clustering-algorithm (2015).
Maeda, R. K. & Karch, F. The ABC of the BX-C: the bithorax complex explained. Development 133, 1413–1422 (2006).
Bartholomew, N. R., Burdett, J. M., VandenBrooks, J. M., Quinlan, M. C. & Call, G. B. Impaired climbing and flight behaviour in Drosophila melanogaster following carbon dioxide anaesthesia. Sci. Rep. 5, 15298 (2015).
Acknowledgements
We thank W. Bender for providing the Fub and Fab-7 mutants, and for discussions; N. Sinnott-Armstrong for help with assembly and programming of the fluid handling robot; D. Kingsley, P. Beachy, M. Fuller, A. Villeneuve, A. Rajpurkar, and T.C. Hung for critical reading of the manuscript; J. Wysocka for the mESCs; and B. Gu for assistance with cell culture. This work was supported by the Searle Scholars Program, Burroughs Wellcome Careers at the Scientific Interface Award, Dale Frey Award, Beckman Young Investigator Program, NIH New Innovator Award DP2 (DGM132935A), and the Packard Fellows Program (A.N.B.). L.J.M. is supported by the Stanford School of Medicine Dean’s Fund, S.E.M. is supported by the Stanford Genome Training Program, A.H. is supported by a Walter V. and Idun Berry Postdoctoral Fellowship, and I.S.C. was supported by a summer training grant from Stanford Bio-X.
Author information
Authors and Affiliations
Contributions
L.J.M. and A.N.B. designed the experiments. L.J.M. collected the data with assistance from S.E.M., I.S.C., C.A.W. and A.N.B. A.H. collected and analysed the mouse data. L.J.M., S.E.M., A.H. and A.N.B. analysed the data and wrote the manuscript with assistance from I.S.C. and C.A.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 ORCA methodology and experimental controls.
a, Schematic of imaging approach. Top, oligo species involved in labelling. To minimize costs, only two oligo sequences carry fluorophore labels: a fiducial oligo labelled with a Cy3 dye and an imaging oligo labelled with a Cy5 dye. Sequence-specific binding is achieved by in situ hybridization of the primary probes to the genomic DNA. Each primary probe consists of a 40-nt targeting region, a 20-nt arm to bind the fiducial oligo, and a 20-nt barcode to bind the readout oligo. Each barcoded region has at least 20 primary probes, to facilitate detection. Other relevant oligo species include a readout oligo for each barcode (Supplementary Table 5) and a strand-displacement oligo for each barcode (Supplementary Table 6). Bottom, data acquisition sequence. The fiducial oligo binds all primary probes and remains bound throughout the experiment. This enables image registration in the downstream image analysis. The readout oligo binds its target barcode ‘1’ and the Cy5 imaging oligo. The now fluorescent barcode ‘1’ is imaged simultaneously with the fiducial Cy3 signal in 3D. The readout oligo ‘1’ is removed by its corresponding strand-displacement oligo, removing the Cy5 signal. This process repeats for the rest of the barcodes. b, Efficient removal of fluorescent signal by strand displacement: top, the fluorescent Cy5 signal from labelled barcode ‘1’ and the corresponding Cy3 signal from the fiducial channel. Bottom, the same region after treatment with the strand-displacement oligo. Centre panels show the Cy5 channel in increased contrast, showing that all the Cy5 fluorescent signal is removed. Notably, the Cy3 image is unchanged. Similar results were obtained in 6,933 cells analysed. c, Violin plots of the error in nanometres along each image axis, determined as the difference between the original measurements and measurements repeated at the end of the experiment. Red line marks the median, black marks the mean. Distribution of errors based on five repeated barcode-position measurements, n = 163,810 repeated barcodes (left). Distribution of errors from measurement of a single barcode’s position where the original and repeated measurements are separated by less than one hour (h), n = 31,506 repeated barcodes (centre). Distribution of errors from measurement of a single barcode’s position when the original and repeated measurements were separated by the entire experiment lasting 96 h n = 40,032 repeated barcodes (right). N.S. indicates no statistically significant increase in the 3D error (P = 0.8, one-sided Wilcoxon test). d, Correlation between the replicates of ORCA experiments with both 10-kb barcode resolution (top) and 3-kb barcode resolution (bottom) when measuring contact frequency (left) and distance (right). Pearson’s correlation coefficient, r, is indicated. e, Pearson’s correlation of contact frequency measured by ORCA and published Hi-C measurements23 for the 10-kb and 3-kb resolution probesets. f, Pearson’s correlation coefficient between the 150-nm cutoff (used in the main figures) and alternative cutoff values from 50 to 500 nm. Pearson’s r was computed using all unique pairwise combinations of all barcodes measured (70 barcodes in the 10-kb resolution experiments and 52 in both the 3-kb and 2-kb resolution experiments). Relative interaction frequencies and corresponding structural boundaries observed in the contact frequency maps have little dependence on the precise value of the cutoff. Alternate cutoffs remain highly correlated to one another over a range of values.
Extended Data Fig. 2 Barcode detection efficiency.
a, The mean detection efficiency of each barcode for all BX-C probesets. Error bars represent s.e.m. n = 8,801 cells, n = 94,300 cells, and n = 15,230 cells for the 10-kb, 3-kb, and 2-kb probesets, respectively. b, Box-and-whisker plots comparing the distribution of detection efficiencies across barcodes between two different embryos. N.S. indicates not statistically significant (two-sided Wilcoxon test, P values indicated). For the boxplots in b–e, the red line marks the median, the upper and lower limits of the boxes mark the interquartile range, and the whiskers extend to the farthest data points within 1.5 times the interquartile range. Notches denote the 95% confidence interval around the median. Sample size (n) is indicated. c, Comparison of the distribution of detection efficiencies between cells of different embryos. d, Comparison of the detection efficiency across barcodes between anterior and posterior cells. e, Comparisons of the detection efficiency across all cells between anterior and posterior cells. Results are shown for the 10-kb, 3-kb and 2-kb probesets as indicated. f, Correlation between normalized distance data for each body segment as shown in Fig. 4 and a modified dataset in which 50% of detected barcodes were removed at random to simulate a high missed detection rate. The correlation coefficients are indicated (Pearson), computed using all unique combinations of 52 barcodes. g, Bar graph comparing the mean correlation between matched segments in the original and downsampled data shown in f compared to the mean inter-segment correlation. The correlation coefficients are indicated (Pearson), computed using all unique combinations of 52 barcodes. All 15 matched segment pairs shown in f are plotted compared to all 210 inter-segment pairs. This illustrates that the difference between the substantially downsampled data and the original measurements are much smaller than the typical differences between segments, demonstrating the robustness of the segment-specific findings to missing data. Error bars denote s.e.m. h, Histogram showing the detection efficiency per cell for each probeset.
Extended Data Fig. 3 STORM analysis of paired Drosophila homologues.
a, Schematic of expected outcomes from STORM if chromosomes are tightly paired and follow a similar trajectory, or are loosely paired and follow distinct trajectories that are within a diffraction-limited distance of one another. b, Representative images from STORM of paired homologues labelled with ORCA barcode 69. Similar images were observed for all barcodes shown in c, d. Centres from 3D Gaussian fitting of the point spread function are represented as plus symbols on spots. Occasional double peaks occur at a frequency similar to that of background non-specific localizations; these may represent stray probes, noise in peak detection of weakly labelled loci, or split homologues. c, d, Quantification of the frequency of single peak detection for 10 kb (c) and 3 kb (d) per barcode. The high level of single peak detection is indicative of the tightly paired homologues for the BX-C. For the boxplots in c, d, the red line marks the median, the upper and lower limits of the boxes mark the interquartile range, and the whiskers extend to the farthest data points within 1.5 times the interquartile range. Number of foci (n) is indicated below each box plot.
Extended Data Fig. 4 ORCA in mouse embryonic stem cells.
a, Raw image of mouse embryonic stem cells with the Sox2 region labelled, showing two distinct homologues per cell. Similar images were observed for all 2,240 cells. b, Distance maps from two single cells, and the corresponding ORCA images. c, Average contact frequency as measured by ORCA (150-nm threshold). The locations of the CTCF ChIP-seq24 peaks, the Sox2 gene, and the ORCA barcodes are indicated. d, Average contact frequency as measured by high resolution Hi-C24. The TAD containing Sox2 is outlined with black lines and the weak corner point (CTCF loop) is marked with a circle. e, Pearson correlation between the data in c and d. Pearson’s correlation and Pearson’s r in e, g were computed using all unique pairwise combinations of all 33 barcodes measured. f, Genome browser view of the Sox2 region, showing the position of the 5-kb barcodes relative to the gene, its regulatory region, and local CTCF peaks. g, Pearson’s correlation coefficients from comparing different choices of the threshold distance for contact frequency as indicated.
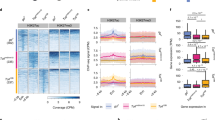
Extended Data Fig. 5 Multiplex smFISH and quantification.
a, Sequentially acquired images from multiplex smFISH and intron-FISH for 18 cytosolic mRNAs and 11 nascent RNAs, respectively, in a laterally cryo-sectioned Drosophila embryo about 11 hpf. b, A dorsolateral cryosection of another embryo, about 11 hpf, showing the positions of all RNAs quantified. The zoomed-in view shows the segmented cells and a higher-resolution depiction of the RNA positions. Bright foci for each mRNA transcript (Fig. 2a) were identified using an automated counting algorithm with a manually defined threshold. The embryo images were segmented to identify cell boundaries and each RNA transcript identified is depicted as a single spot, colour-coded by RNA species. Note, Abd-B has two differentially expressed isoforms (known as m and r) which are transcribed from different promoters. Similar results to those shown were obtained for 15 embryos.
Extended Data Fig. 6 Distinct spatial organization and chromatin structure per cell type.
a, t-SNE 2D projection of the RNA expression data, clustered with DBSCAN (see Methods). Annotations identified by manual inspection are indicated by matching colours and numbers (labelled on the right). This number/colour legend is used for all panels. b, The relative embryonic spatial positions of cells in each group shown for three embryos. c, Distance maps from ORCA for each assigned group. d, Normalized distance maps (observed minus expected distance) for each group. The expected distance accounts for the polymer nature of DNA, whereby sequences closer in linear position along the genome are expected to be closer together in 3D space. The expected distance was calculated by fitting a power law to the distribution of 3D separation distances between barcodes as a function of their linear separation for all data in all cells.
Extended Data Fig. 7 Enhancer-promoter interactions and single cell examples.
a, Violin plots showing the distance between each indicated enhancer and its cognate promoter in cells in which nascent transcripts were detected or absent. The average distances are marked by plus symbols. P values from a two-sided KS test between the distributions (PKS) and from a two-sided Wilcoxon test (PW) are shown. The total number of cells, n, in each distribution is indicated. The enhancers from each genetic domain (two from each of the largest domains) are labelled by their classical names. The relative positions of enhancers and promoters are shown below. Note that most cells from the silent state come from anterior regions where the BX-C is repressed and compacted by Polycomb. Despite this compaction of the repressed state and relative decompaction of the active state, all enhancer–promoter pairs are on average slightly closer when active. A more substantial difference may be expected for domains in which the silent state is not largely attributable to Polycomb activity. b, Two example distance maps from single cells for each embryonic segment as indicated. Population-average normalized maps as in Fig. 4 are shown below for comparison.
Extended Data Fig. 8 Automated identification of TAD boundaries from insulation score.
Relative distance maps for each segment, plotted at 3-kb resolution. TAD boundaries (dotted black lines) were identified by an automated algorithm based on computation of the insulation score (plotted below each graph) and application of a threshold. A threshold score of 0.4 was used to call TAD boundaries, which agree with the boundaries detected by manual inspection and annotated as in Fig. 4. The insulation score for each segment was computed as follows: for each barcode, we first measured a score for the upstream region by computing the fraction of red bins (normalized distance <0) in a block of 6 × 6 bins and a similar score for the downstream block and for the interblock region. To compute the insulation score, we then took the average scores for the two blocks minus the score of the interblock.
Extended Data Fig. 9 Comparison of 3-kb segment-specific datasets.
a, Normalized distance maps from dataset 1, embryos 10–12 hpf. b, Normalized distance maps from dataset 2, embryos 8–10 hpf, showing similar patterns to dataset 1. All maps are normalized relative to cells from the head segments, as in Fig. 4. c, Pearson correlation between datasets 1 and 2 by segment. Note that after normalization, organization in the head and anterior thorax is apparently random, and thus uncorrelated between datasets. The systematic features that appear in posterior segments, in which regulatory sequences of the BX-C are active, are similar between datasets. d, Pearson correlation of non-normalized distance data. In c and d, values from all unique combinations of the 52 barcodes were used to compute the correlation.
Extended Data Fig. 10 Properties of Fub mutant embryos.
a, Expression of abd-A and Abd-B in wild-type (WT) and Fub mutant embryos. Note that abd-A expression expands to T3 in mutant embryos, compared to wild-type embryos. b, ORCA data at 10-kb resolution for the 330-kb BX-C region, normalized as in Fig. 4, for cells from segment A1 in wild-type and Fub mutant embryos. Predicted TADs are marked by black lines. The dotted circle over the Fub mutant distance map highlights aberrant interactions. The extent of H3K27me3 measured in A134 in wild-type embryos and the predicted extent of H3K27me3 in Fub mutants are shown in purple. Predicted H3K27me3 is based on the activation of abd-A observed in the mutant and previous descriptions of phenotypes of genetic border deletions in Drosophila25,26,27. The region deleted in Fub mutants is marked by red dotted lines. c, Fecundity of wild-type, homozygous Fab-7 and balanced Fub lines, plotted as the number of viable adults produced by eight fertilized females in a 2-day window. d, The height that each Drosophila larva, represented as a dot, climbed before pupation. Average height is indicated on the x-axis. e, Comparison of flight ability between wild-type and Fub/MKRS lines, using the metric developed by Bartholomew et al.63. The metric reports the fraction of adult Drosophila that re-land on the sides of the chamber rather than falling to the bottom, after being displaced by light tapping. Each dot represents a separate flight trial. Red line represents the median, boxes mark the upper and lower quartiles, whiskers extend to the farthest data points within 1.5 times the interquartile range.
Supplementary information
Supplementary Data Table 1
Bed file containing annotated positions of all readout probes, the 10 genetic domains, the previously identified enhancers, the previously identified insulators, and the ChIP-seq data for CTCF, CP190 and RAD21 proteins, throughout the regions examined here. This data can also be viewed through the following link: https://tinyurl.com/y7wf8t6x.
Supplementary Data Table 2
This file contains Barcode Sequences, as an Excel file.
Supplementary Data Table 3
This file contains Primer Sequences, as an Excel file.
Supplementary Data Table 4
This file contains Oligopaint Probe Sequences, as a Fasta file.
Supplementary Data Table 5
This file contains Readout Probe Sequences, as an Excel file.
Supplementary Data Table 6
This file contains Strand-displacement Oligo Sequences, as an Excel file.
Rights and permissions
About this article
Cite this article
Mateo, L.J., Murphy, S.E., Hafner, A. et al. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568, 49–54 (2019). https://doi.org/10.1038/s41586-019-1035-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1035-4
This article is cited by
-
pyHiM: a new open-source, multi-platform software package for spatial genomics based on multiplexed DNA-FISH imaging
Genome Biology (2024)
-
Multiplex DNA fluorescence in situ hybridization to analyze maternal vs. paternal C. elegans chromosomes
Genome Biology (2024)
-
Simultaneous single-cell three-dimensional genome and gene expression profiling uncovers dynamic enhancer connectivity underlying olfactory receptor choice
Nature Methods (2024)
-
Boundary stacking interactions enable cross-TAD enhancer–promoter communication during limb development
Nature Genetics (2024)
-
Real-time single-molecule imaging of transcriptional regulatory networks in living cells
Nature Reviews Genetics (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.