Abstract
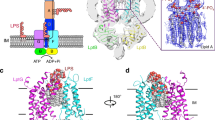
In Gram-negative bacteria, lipopolysaccharide is essential for outer membrane formation and antibiotic resistance. The seven lipopolysaccharide transport (Lpt) proteins A–G move lipopolysaccharide from the inner to the outer membrane. The ATP-binding cassette transporter LptB2FG, which tightly associates with LptC, extracts lipopolysaccharide out of the inner membrane. The mechanism of the LptB2FG–LptC complex (LptB2FGC) and the role of LptC in lipopolysaccharide transport are poorly understood. Here we characterize the structures of LptB2FG and LptB2FGC in nucleotide-free and vanadate-trapped states, using single-particle cryo-electron microscopy. These structures resolve the bound lipopolysaccharide, reveal transporter–lipopolysaccharide interactions with side-chain details and uncover how the capture and extrusion of lipopolysaccharide are coupled to conformational rearrangements of LptB2FGC. LptC inserts its transmembrane helix between the two transmembrane domains of LptB2FG, which represents a previously unknown regulatory mechanism for ATP-binding cassette transporters. Our results suggest a role for LptC in achieving efficient lipopolysaccharide transport, by coordinating the action of LptB2FG in the inner membrane and Lpt protein interactions in the periplasm.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Seven 3D cryo-EM density maps of E. coli LptB2FG and LptB2FGC in nanodiscs have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-9118 (nucleotide-free LptB2FG), EMD-9125 (nucleotide-free LptB2FGC, final map), EMD-9128 (nucleotide-free LptB2FGC, map with clear LPS density), EMD-9129 (nucleotide-free LptB2FGC, βJRlong map), EMD-9130 (nucleotide-free LptB2FGC, βJRshort map), EMD-9124 (vanadate-trapped LptB2FG) and EMD-9126 (vanadate-trapped LptB2FGC). Four atomic coordinates for the atomic models have been deposited in the PDB under accession numbers 6MHU (nucleotide-free LptB2FG), 6MI7 (nucleotide-free LptB2FGC), 6MHZ (vanadate-trapped LptB2FG) and 6MI8 (vanadate-trapped LptB2FGC). Any other relevant data are available from the corresponding author upon reasonable request.
References
Boucher, H. W. et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12 (2009).
Raetz, C. R. & Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 (2002).
Raetz, C. R., Reynolds, C. M., Trent, M. S. & Bishop, R. E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 (2007).
Whitfield, C. & Trent, M. S. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 (2014).
Beutler, B. & Rietschel, E. T. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 3, 169–176 (2003).
Ruiz, N., Kahne, D. & Silhavy, T. J. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat. Rev. Microbiol. 7, 677–683 (2009).
May, J. M., Sherman, D. J., Simpson, B. W., Ruiz, N. & Kahne, D. Lipopolysaccharide transport to the cell surface: periplasmic transport and assembly into the outer membrane. Phil. Trans. R. Soc. Lond. B 370, 20150027 (2015).
Simpson, B. W., May, J. M., Sherman, D. J., Kahne, D. & Ruiz, N. Lipopolysaccharide transport to the cell surface: biosynthesis and extraction from the inner membrane. Phil. Trans. R. Soc. Lond. B 370, 20150029 (2015).
Okuda, S., Sherman, D. J., Silhavy, T. J., Ruiz, N. & Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345 (2016).
Sherman, D. J. et al. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801 (2018).
Suits, M. D., Sperandeo, P., Dehò, G., Polissi, A. & Jia, Z. Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J. Mol. Biol. 380, 476–488 (2008).
Sherman, D. J. et al. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc. Natl Acad. Sci. USA 111, 4982–4987 (2014).
Tran, A. X., Dong, C. & Whitfield, C. Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 (2010); corrigendum 292, 18731 (2017).
Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511, 52–56 (2014).
Qiao, S., Luo, Q., Zhao, Y., Zhang, X. C. & Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511, 108–111 (2014).
Dong, H., Zhang, Z., Tang, X., Paterson, N. G. & Dong, C. Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG. Nat. Commun. 8, 222 (2017).
Luo, Q. et al. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol. 24, 469–474 (2017).
Laguri, C. et al. Interaction of lipopolysaccharides at intermolecular sites of the periplasmic Lpt transport assembly. Sci. Rep. 7, 9715 (2017).
Thomas, C. & Tampé, R. Multifaceted structures and mechanisms of ABC transport systems in health and disease. Curr. Opin. Struct. Biol. 51, 116–128 (2018).
Narita, S. & Tokuda, H. Biochemical characterization of an ABC transporter LptBFGC complex required for the outer membrane sorting of lipopolysaccharides. FEBS Lett. 583, 2160–2164 (2009).
Freinkman, E., Okuda, S., Ruiz, N. & Kahne, D. Regulated assembly of the transenvelope protein complex required for lipopolysaccharide export. Biochemistry 51, 4800–4806 (2012).
Villa, R. et al. The Escherichia coli Lpt transenvelope protein complex for lipopolysaccharide export is assembled via conserved structurally homologous domains. J. Bacteriol. 195, 1100–1108 (2013).
Martorana, A. M. et al. Functional interaction between the cytoplasmic ABC protein LptB and the inner membrane LptC protein, components of the lipopolysaccharide transport machinery in Escherichia coli. J. Bacteriol. 198, 2192–2203 (2016).
Schultz, K. M. & Klug, C. S. Characterization of and lipopolysaccharide binding to the E. coli LptC protein dimer. Protein Sci. 27, 381–389 (2018).
Okuda, S., Freinkman, E. & Kahne, D. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217 (2012).
Jeong, H. et al. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J. Mol. Biol. 394, 644–652 (2009).
Bertani, B. R., Taylor, R. J., Nagy, E., Kahne, D. & Ruiz, N. A cluster of residues in the lipopolysaccharide exporter that selects substrate variants for transport to the outer membrane. Mol. Microbiol. 109, 541–554 (2018).
Locher, K. P. Mechanistic diversity in ATP-binding cassette (ABC) transporters. Nat. Struct. Mol. Biol. 23, 487–493 (2016).
Dong, H., Tang, X., Zhang, Z. & Dong, C. Structural insight into lipopolysaccharide transport from the Gram-negative bacterial inner membrane to the outer membrane. Biochim. Biophys. Acta 1862, 1461–1467 (2017).
Doerrler, W. T. & Raetz, C. R. ATPase activity of the MsbA lipid flippase of Escherichia coli. J. Biol. Chem. 277, 36697–36705 (2002).
Oldham, M. L. & Chen, J. Snapshots of the maltose transporter during ATP hydrolysis. Proc. Natl Acad. Sci. USA 108, 15152–15156 (2011).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Mi, W. et al. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 549, 233–237 (2017).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Ru, H. et al. Molecular mechanism of V(D)J recombination from synaptic RAG1–RAG2 complex structures. Cell 163, 1138–1152 (2015).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015).
Swint-Kruse, L. & Brown, C. S. Resmap: automated representation of macromolecular interfaces as two-dimensional networks. Bioinformatics 21, 3327–3328 (2005).
Lyumkis, D., Brilot, A. F., Theobald, D. L. & Grigorieff, N. Likelihood-based classification of cryo-EM images using FREALIGN. J. Struct. Biol. 183, 377–388 (2013).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Kim, H. M. et al. Crystal structure of the TLR4–MD-2 complex with bound endotoxin antagonist eritoran. Cell 130, 906–917 (2007).
van Aalten, D. M. et al. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 10, 255–262 (1996).
Kyte, J. & Doolittle, R. F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Acknowledgements
We thank Z. Li and M. Chambers at HMS and C. Xu and K. Song at the UMass cryo-EM facility for electron microscopy technical support; T. Silhavy for providing the lptFG-deletion strain NR1113; T. Walz, T. Rapoport, T. Walther and W. Harper for critical reading of the manuscript; and members of the Liao group for helpful discussions and comments on the manuscript. This work was supported by NIH grant R01GM122797 to M.L.
Reviewer information
Nature thanks Russell Bishop, Alessandra Polissi, Robert Tampé and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
M.L. conceived the project. Y.L. performed cloning, expression, purification, nanodisc incorporation and functional assays. Y.L. and B.J.O. performed ATPase activity measurements and screened samples using negative-stain electron microscopy. B.J.O. prepared cryo-EM grids, collected and processed cryo-EM data and built the atomic models. All authors analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Purification and functional characterization of LptB2FG and LptB2CFG in DDM and in nanodiscs.
a, Gel-filtration chromatography profile of LptB2FG in DDM. b, Gel-filtration chromatography profile of LptB2FG in nanodiscs. c, Coomassie-blue-stained SDS–PAGE gel of purified LptB2FG in DDM and in nanodiscs. Individual protein components of the complex are labelled. d, Gel-filtration chromatography profile of LptB2FGC in DDM. e, Gel-filtration chromatography profile of LptB2FGC in nanodiscs. f, Coomassie-blue-stained SDS–PAGE gel of purified LptB2FGC in DDM and in nanodiscs. Individual protein components of the complex are labelled. The experiments in a–f were repeated three times independently with similar results. g, ATPase activity of LptB2FG and LptB2FGC in DDM and in nanodiscs. Each point represents mean ± s.d. of three separate measurements. h, Vanadate-concentration-dependent inhibition of the ATPase activity of nanodisc-reconstituted LptB2FG and LptB2FGC. Each point represents mean ± s.d. of three separate measurements. For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 2 Image processing for the cryo-EM data of nucleotide-free LptB2FG in nanodiscs.
a, Representative cryo-EM image of nucleotide-free LptB2FG in nanodiscs. b, Two-dimensional class averages of cryo-EM particle images. c, Three-dimensional classification and refinement of cryo-EM particle images. After the first round of 3D classification, all particles classified into the two best classes (no. 3 and no. 6) in the final 5 iterations (indicated as ‘5 cycles’) were kept for further processing. Three-dimensional classification focusing on the TMD was used to obtain the final cryo-EM map. LPS density is indicated with a green circle. Electron microscopy data collection and 2D classification were performed once.
Extended Data Fig. 3 Single-particle cryo-EM analysis of nucleotide-free LptB2FG in nanodiscs.
a, Local resolution of the final cryo-EM map of nucleotide-free LptB2FG. b, FSC curves: gold-standard FSC curve between the two half maps with indicated resolution at FSC = 0.143 (red); FSC curve between the atomic model and the final map with indicated resolution at FSC = 0.5 (blue); FSC curve between half map 1 (orange) or half map 2 (green) and the atomic model refined against half map 1. c, Cutaway views of angular distribution of particle images included in the final 3D reconstruction. d, Surface view and sectional view of the cryo-EM map of nucleotide-free LptB2FG filtered to 6 Å resolution to show the lipid nanodisc, overall arrangement of transmembrane helices, β-jellyroll domains and LPS (left). Slices through the cryo-EM map at the indicated planes. Arrowhead and arrows indicate the inner core and the phosphorylated glucosamines, respectively. Individual transmembrane helices are numbered in the bottom slice view. This analysis was performed once. e, Front and back TMD interfaces formed by the TM1 and TM5 helices from LptF and LptG, coloured in orange and blue, respectively. LPS is shown as spheres. f, Cryo-EM densities superimposed with the atomic model for individual transmembrane helices in the nucleotide-free LptB2FG. g, Cryo-EM densities superimposed with the atomic model for selected regions of the NBDs (LptB), which demonstrates the clear separation of the β-strands and side-chain densities.
Extended Data Fig. 4 Hydrophobic and electrostatic interactions between LPS and LptB2FG.
a, Surface and sectional views of hydrophobic surface representation of nucleotide-free LptB2FG, showing hydrophobic (orange) and hydrophilic (blue) areas. LPS is shown as green sticks. Right, view perpendicular to the membrane plane, with the transmembrane helices and several acyl-chain-interacting side chains shown as ribbons and sticks, respectively. b, Surface and sectional views of electrostatic surface representation of nucleotide-free LptB2FG, showing areas of positive (blue) and negative (red) charge. LPS is shown as green sticks. c, Sectional views from the periplasm at the four different planes indicated in the right panel in a, showing electrostatic and hydrophobic interactions of LPS with LptF and LptG. Cryo-EM density (grey surface) is superimposed with the atomic model. Side chains that interact with 1-PO4, 4′-PO4 and the acyl chains of LPS are labelled. d, Side views of the same regions in the 4 Å resolution cryo-EM map (left) and the 2Fo − Fc electron density map for the 3.46 Å resolution crystal structure (PDB: 5L75) (right). Electrostatic interactions with the 1-PO4 and 4′-PO4 groups stabilize the side chains of R133 and R136 in LptG, and K30 and R33 in LptF.
Extended Data Fig. 5 Image processing for the cryo-EM data of nucleotide-free LptB2FGC in nanodiscs.
Different subsets of particle images were selected from different classification schemes to produce four refined cryo-EM maps: final LptB2FGC map at 4.2 Å resolution, LptB2FGC map with clear LPS density at 4.4 Å resolution, βJRlong LptB2FGC map at 4.8 Å resolution and βJRshort LptB2FGC map at 5.9 Å resolution. After the first round of 3D classification, all particles that were classified into the two best classes (no. 4 and no. 5) in the final five iterations (indicated as ‘5 cycles’) were kept for further processing.
Extended Data Fig. 6 Single-particle cryo-EM analysis of nucleotide-free LptB2FGC in nanodiscs.
a, Representative cryo-EM image of nucleotide-free LptB2FGC in nanodiscs. b, Two-dimensional class averages of cryo-EM particle images. c, Local resolution of the final cryo-EM map of nucleotide-free LptB2FGC. d, FSC curves: gold-standard FSC curve between the two half maps with indicated resolution at FSC = 0.143 (red); FSC curve between the atomic model and the final map with indicated resolution at FSC = 0.5 (blue); FSC between half map 1 (orange) or half map 2 (green) and the atomic model refined against half map 1. e, Cutaway views of angular distribution of particles included in the final 3D reconstruction. f, Cryo-EM densities superimposed with the atomic model for individual transmembrane helices in the nucleotide-free LptB2FGC. g, Cryo-EM density superimposed with the atomic model for a lipid molecule (POPG in green) and surrounding TMLptC, TM5LptF and TM6LptF. This density was modelled as a POPG molecule, because POPG was used for nanodisc reconstitution and is also abundant in the inner membrane of E. coli. Electron microscopy data collection and 2D classification were performed once.
Extended Data Fig. 7 Analysis of the cryo-EM structure of nucleotide-free LptB2FGC.
a, Local resolution of the LptB2FGC map with clear LPS density. b, Gold-standard FSC curves between the two half maps for the three cryo-EM structures of nucleotide-free LptB2FGC. c, Sectional views of the final LptB2FGC map (4.2 Å resolution), the LptB2FGC map with clear LPS density (4.4 Å resolution) and the final LptB2FG map (4.0 Å resolution), all low-pass-filtered to 6.0 Å resolution to compare the density of the phosphorylated glucosamines of the bound LPS. d, Sectional side view (left) and top-down views at two different levels (right) of the LptB2FGC map with clear LPS density (grey), superimposed with the atomic model. LPS density is coloured in green. The four LPS-interacting residues are labelled. e, Sectional front views of the atomic models of LptB2FGC and LptB2FG that were aligned using the two LptB subunits as in g. LPS molecules are shown as green sticks. The two dashed lines indicate the heights at the level of the oxygen atom (red asterisk) in the ether bond that connects the two glucosamines. The distance between the positions of this oxygen atom in the LptB2FGC and LptB2FG structures is 6 Å. f, Functional analysis of R33 of LptF in the bacterial strain NR1113, which is depleted of lptF and lptG. All of the complementation assays were repeated three times independently with similar results, and one representative result is shown. g, Three perpendicular views of the superimposed atomic models of LptB2FG (grey) and LptB2FGC (coloured as in Fig. 3a). Two structures are aligned using the two LptB subunits. h, Views from the periplasm of the LPS-binding pocket in the structures of LptB2FGC (top) and LptB2FG (bottom), shown as ribbon diagram (left) and electrostatic surface (right). The residues that mediate electrostatic interactions with LPS in either LptB2FG or LptB2FGC are labelled.
Extended Data Fig. 8 Image processing for the cryo-EM data of nanodisc-embedded LptB2FG and LptB2FGC with vanadate.
a, Representative cryo-EM image of vanadate-trapped LptB2FG in nanodiscs. b, Two-dimensional class averages of cryo-EM particle images of vanadate-trapped LptB2FG in nanodiscs. c, Two-dimensional class averages of cryo-EM particle images of vanadate-trapped LptB2FGC in nanodiscs. d, Image processing workflow of vanadate-trapped LptB2FG. e, Image processing workflow of vanadate-trapped LptB2FGC. After the first round of 3D classification, all particles that were classified into the best classes in the final five iterations (indicated as ‘5 cycles’) were kept for further processing. Electron microscopy data collection and 2D classification were performed once.
Extended Data Fig. 9 Single-particle cryo-EM analysis of nanodisc-embedded LptB2FG and LptB2FGC with vanadate.
a, Local resolution of the cryo-EM map of vanadate-trapped LptB2FG. b, FSC curves: gold-standard FSC curve between the two half maps with indicated resolution at FSC = 0.143 (red); FSC curve between the atomic model and the final map with indicated resolution at FSC = 0.5 (blue); FSC between half map 1 (orange) or half map 2 (green) and the atomic model refined against half map 1. c, Cutaway views of angular distribution of particles included in the final 3D reconstructions of vanadate-trapped LptB2FG (top) and LptB2FGC (bottom). d, As in a, except for vanadate-trapped LptB2FGC. e, As in b, except for vanadate-trapped LptB2FGC. f, Representative 2D class averages (top) and slices of 3D reconstructions (bottom) of nucleotide-free (left) and vanadate-trapped (right) LptB2FG. LPS density is indicated with a green arrow. These two slices are the same as the bottom slice in Extended Data Fig. 3d and the bottom slice in h. g, Cryo-EM densities with the atomic models for individual transmembrane helices in the vanadate-trapped LptB2FG. h, Cryo-EM map of vanadate-trapped LptB2FG filtered to 6 Å resolution to show the overall arrangement of the transmembrane helices and LptB subunits. Slices through the 3D map at the indicated planes show the positions of individual transmembrane helices and the collapse of the inner cavity (left). Overlays of the TMDs of LptF or LptG in the nucleotide-free (grey) and vanadate-trapped (blue and orange) LptB2FG show only small differences within each TMD (right). Red arrow indicates the bending of TM1LptF upon nucleotide binding. i, Cryo-EM densities for the ADP–vanadate complexes trapped at the two ATP-binding sites between the LptB subunits in LptB2FGC and LptB2FG. The Walker A and signature motifs are coloured in grey and red, respectively. j, Proposed model for LptB2FGC-driven LPS extraction. The Lpt proteins are coloured as in Fig. 3a. Three β-jellyroll domains, the TMLptC and the LptC linker are labelled. LPS is depicted as a cartoon model of lipid A with the inner core. The TMLptC and LptC linker are shown as dashed lines in steps 3 to 5 to indicate their increased mobility. The ATP molecule, before or after hydrolysis, is indicated as a red diamond sandwiched between the two LptB subunits. Additional cycles of LPS extraction are between steps 4 and 5. See ‘Discussion’ in the main text for description of proposed LPS-extraction cycle. The analyses in f and h were performed once.
Supplementary information
Supplementary Figure 1
This file contains the uncropped scans with size marker indications for the Coomassie blue-stained SDS-PAGE gels.
Supplementary Video 1 | LPS density with the atomic model
The animation shows a 360° view of the atomic model of LPS superimposed with the EM density from the cryo-EM map of nucleotide-free LptB2FG. Density is present for all six acyl chains, phosphorylated glucosamines, and the inner core.
Supplementary Video 2 | Conformational rearrangement of the amino acids that mediate electrostatic interactions with LPS
The amination shows a morph between the cryo-EM structures of nucleotide-free LptB2FGC and LptB2FG, viewed from the periplasm at the LPS-binding pocket. LPS and TMC are omitted. The periplasmic end of TM1G containing Lys40 and Lys41 is disordered in the structure of LptB2FGC and not shown. Side chains are shown only for the LPS-interacting residues.
Supplementary Video 3 | Conformational changes of LptB2FG from nucleotide-free to vanadate-trapped state
The animation shows a morph of the structures of the TMDs and NBDs in LptB2FG before and after trapping with vanadate, viewed from the periplasm. LptB subunits are colored green and yellow, LptF is colored orange, and LptG is colored blue. The animation begins with nucleotide-free LptB2FG and progresses to the vanadate-trapped conformation.
Rights and permissions
About this article
Cite this article
Li, Y., Orlando, B.J. & Liao, M. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567, 486–490 (2019). https://doi.org/10.1038/s41586-019-1025-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-1025-6
This article is cited by
-
Molecular insights into capsular polysaccharide secretion
Nature (2024)
-
A new antibiotic traps lipopolysaccharide in its intermembrane transporter
Nature (2024)
-
Macrocyclic peptides: up-and-coming weapons to combat antimicrobial resistance
Signal Transduction and Targeted Therapy (2024)
-
Conformational Investigation of the Asymmetric Periplasmic Domains of E. coli LptB2FGC Using SDSL CW EPR Spectroscopy
Applied Magnetic Resonance (2024)
-
Alternating L4 loop architecture of the bacterial polysaccharide co-polymerase WzzE
Communications Biology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.