Abstract
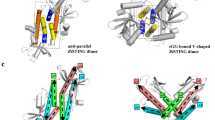
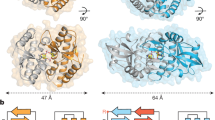
Infections by pathogens that contain DNA trigger the production of type-I interferons and inflammatory cytokines through cyclic GMP–AMP synthase, which produces 2′3′-cyclic GMP–AMP (cGAMP) that binds to and activates stimulator of interferon genes (STING; also known as TMEM173, MITA, ERIS and MPYS)1,2,3,4,5,6,7,8. STING is an endoplasmic-reticulum membrane protein that contains four transmembrane helices followed by a cytoplasmic ligand-binding and signalling domain9,10,11,12,13. The cytoplasmic domain of STING forms a dimer, which undergoes a conformational change upon binding to cGAMP9,14. However, it remains unclear how this conformational change leads to STING activation. Here we present cryo-electron microscopy structures of full-length STING from human and chicken in the inactive dimeric state (about 80 kDa in size), as well as cGAMP-bound chicken STING in both the dimeric and tetrameric states. The structures show that the transmembrane and cytoplasmic regions interact to form an integrated, domain-swapped dimeric assembly. Closure of the ligand-binding domain, induced by cGAMP, leads to a 180° rotation of the ligand-binding domain relative to the transmembrane domain. This rotation is coupled to a conformational change in a loop on the side of the ligand-binding-domain dimer, which leads to the formation of the STING tetramer and higher-order oligomers through side-by-side packing. This model of STING oligomerization and activation is supported by our structure-based mutational analyses.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The cryo-EM maps generated in this study have been deposited in the Electron Microscopy Data Bank (EMDB) under the accession numbers EMD-0502 (human STING, apo dimer), EMD-0503 (chicken STING, apo dimer), EMD-0504 (chicken STING, cGAMP-bound dimer) and EMD-0505 (chicken STING, cGAMP-bound tetramer). The atomic coordinates have been deposited in the PDB under the accession numbers 6NT5 (human STING, apo dimer), 6NT6 (chicken STING, apo dimer), 6NT7 (chicken STING, cGAMP bound dimer) and 6NT8 (chicken STING, cGAMP bound tetramer).
References
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Wu, J. et al. Cyclic GMP–AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339, 826–830 (2013).
Cai, X., Chiu, Y. H. & Chen, Z. J. The cGAS–cGAMP–STING pathway of cytosolic DNA sensing and signaling. Mol. Cell 54, 289–296 (2014).
Ishikawa, H. & Barber, G. N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455, 674–678 (2008).
Zhong, B. et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29, 538–550 (2008).
Burdette, D. L. et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011).
Jin, L. et al. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol. Cell. Biol. 28, 5014–5026 (2008).
Sun, W. et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl Acad. Sci. USA 106, 8653–8658 (2009).
Zhang, X. et al. Cyclic GMP–AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING. Mol. Cell 51, 226–235 (2013).
Shang, G. et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 19, 725–727 (2012).
Huang, Y. H., Liu, X. Y., Du, X. X., Jiang, Z. F. & Su, X. D. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 19, 728–730 (2012).
Ouyang, S. et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36, 1073–1086 (2012).
Shu, C., Yi, G., Watts, T., Kao, C. C. & Li, P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 19, 722–724 (2012).
Gao, P. et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 154, 748–762 (2013).
Kranzusch, P. J. et al. Ancient origin of cGAS–STING reveals mechanism of universal 2′,3′ cGAMP signaling. Mol. Cell 59, 891–903 (2015).
Zhang, H. et al. Rat and human STINGs profile similarly towards anticancer/antiviral compounds. Sci. Rep. 5, 18035 (2015).
Chen, Q., Sun, L. & Chen, Z. J. Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol. 17, 1142–1149 (2016).
Li, T. & Chen, Z. J. The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018).
Bai, X. C., McMullan, G. & Scheres, S. H. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 40, 49–57 (2015).
Danev, R. & Baumeister, W. Cryo-EM single particle analysis with the Volta phase plate. eLife 5, e13046 (2016).
Yin, Q. et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46, 735–745 (2012).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Li, Z. et al. PPM1A regulates antiviral signaling by antagonizing TBK1-mediated STING phosphorylation and aggregation. PLoS Pathog. 11, e1004783 (2015).
Tanaka, Y. & Chen, Z. J. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci. Signal. 5, ra20 (2012).
Liu, Y. et al. Activated STING in a vascular and pulmonary syndrome. N. Engl. J. Med. 371, 507–518 (2014).
Jeremiah, N. et al. Inherited STING-activating mutation underlies a familial inflammatory syndrome with lupus-like manifestations. J. Clin. Invest. 124, 5516–5520 (2014).
Melki, I. et al. Disease-associated mutations identify a novel region in human STING necessary for the control of type I interferon signaling. J. Allergy Clin. Immunol. 140, 543–552.e545 (2017).
Shi, H., Wu, J., Chen, Z. J. & Chen, C. Molecular basis for the specific recognition of the metazoan cyclic GMP–AMP by the innate immune adaptor protein STING. Proc. Natl Acad. Sci. USA 112, 8947–8952 (2015).
Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature (2019).
Mukai, K. et al. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7, 11932 (2016).
Haag, S. M. et al. Targeting STING with covalent small-molecule inhibitors. Nature 559, 269–273 (2018).
Morales-Perez, C. L., Noviello, C. M. & Hibbs, R. E. Manipulation of subunit stoichiometry in heteromeric membrane proteins. Structure 24, 797–805 (2016).
Dukkipati, A., Park, H. H., Waghray, D., Fischer, S. & Garcia, K. C. BacMam system for high-level expression of recombinant soluble and membrane glycoproteins for structural studies. Protein Expr. Purif. 62, 160–170 (2008).
Lu, D. et al. Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism. Mol. Microbiol. 92, 1092–1112 (2014).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zhang, K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016).
Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Bai, X. C., Rajendra, E., Yang, G., Shi, Y. & Scheres, S. H. Sampling the conformational space of the catalytic subunit of human γ-secretase. eLife 4, e11182 (2015).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We thank Hongtao Yu and Ryan Hibbs for sharing intruments and reagents, and Xiang Gui for his contributions on the analysis of some STING mutants. Cryo-EM data were collected at the University of Texas Southwestern Medical Center (UTSW) Cryo-Electron Microscopy Facility, which is funded by the Cancer Prevention and Research Institute of Texas (CPRIT) Core Facility Support Award RP170644. We thank D. Nicastro for facility access and data acquisition. This work is supported in part by the Howard Hughes Medical Institute (Z.J.C.), grants from the National Institutes of Health (GM088197 and R35GM130289 to X.Z.), grants from the Welch foundation (I-1389 to Z.J.C.; I-1702 to X.Z.; I-1944 to X.-c.B.), grants from CPRIT (RP150498 to Z.J.C.; RP160082 to X.-c.B.). X.-c.B. and X.Z. are Virginia Murchison Linthicum Scholars in Medical Research at UTSW. Z.J.C. is an investigator of Howard Hughes Medical Institute.
Reviewer information
Nature thanks Andrea Ablasser, Philip Kranzusch and Osamu Nureki for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
All authors participated in research design, data analyses and manuscript preparation; C.Z. and G.S prepared the STING protein samples for cryo-EM; X.-c.B., G.S. and X.Z. performed data acquisition, image processing, structure determination and analyses; and C.Z. did the functional assays under the supervision of Z.J.C.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Flow chart of cryo-EM image processing for chicken STING in the apo state.
a, Representative micrograph. b, Representative 2D classes. c, Final reconstruction with colours based on local resolution. d, Gold-standard FSC curve of the final 3D reconstruction. e, Image processing procedure.
Extended Data Fig. 2 Flow chart of cryo-EM image processing for human STING in the apo state.
a, Representative micrograph. b, Representative 2D classes. c, Final reconstruction with colours based on local resolution. d, Gold-standard FSC curve of the final 3D reconstruction. e, Image processing procedure.
Extended Data Fig. 3 Structure of full-length chicken STING in the apo state.
a, Side view of the cryo-EM 3D reconstruction. The two subunits in the dimer are coloured in yellow and green. b, Cartoon representation of the structure in two orthogonal side views. c, Cartoon representation of the transmembrane domain dimer in the top view, from the cytosolic side. d, Interactions between the N-terminal segment and the body of chicken STING. e, Sequence alignment of STING from human, mouse and chicken (denoted by h-, m- and ch- prefixes, respectively). Secondary structure assignments are based on the structures. Residue numbers of human and chicken STING are shown above and below the aligned sequences, respectively.
Extended Data Fig. 4 Flow chart of cryo-EM image processing for cGAMP-bound chicken STING.
a, Representative micrograph. b, Representative 2D classes of the dimer (two top panels) and tetramer (bottom panel). c, e, Final reconstructions of the dimer and tetramer, respectively, coloured on the basis of local resolution. d, f, Gold-standard FSC curves of the final 3D reconstructions of the dimer and tetramer. g, Image processing procedure.
Extended Data Fig. 5 Additional mutational analyses of the transmembrane–LBD connector of human STING.
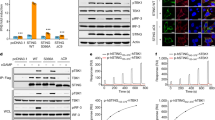
a, c, Effects of mutations in the connector and LBDα1 on cGAMP-stimulated IFNβ expression. Data are mean ± s.d. and representative of three biological replicates. b, d, Effects of STING mutations on phosphorylation of STING, TBK1 and IRF3. The result is representative of two independent experiments. The analyses for gene expression in a, c and phosphorylation in b, d were carried out in the same manner as in Fig. 2c and in Fig. 2d, respectively.
Extended Data Fig. 6 Tetramer of full-length STING.
a, Rigid docking of the atomic model of the cGAMP-bound full-length chicken STING dimer (green and yellow) to the 3D reconstruction of the tetramer (grey). b, Left, tetrameric model of chicken STING in the apo state. The model is generated by superposition of two chicken STING inactive dimers on the active tetramer, on the basis of the transmembrane domain. Right, expanded view of the LBDα2–LBDα3 loop at the tetramer interface. The same loop in the active tetramer structure is shown for comparison; arrows indicate conformational differences between the two states. c, Model of full-length human STING tetramer. The model is constructed by superimposing two inactive human STING dimers on the active chicken STING tetramer, on the basis of the transmembrane domain. Right, top panel, packing between Phe153 in the connector loop and the LBDα2–LBDα3 loop. Right, bottom panel, Cys88 and Cys91—which have been shown to be palmitoylated—are highlighted. d, Cys206, Arg281 and Arg284—alterations of which cause constitutive activation of human STING—are located near the LBDα2–LBDα3 loop that forms the tetramer interface. These residues are highlighted in the tetramer model of the human STING LBD bound to cGAMP (based on PDB code 4KSY, as shown in Fig. 4c).
Extended Data Fig. 7 Sample density maps.
a–c, Sample density maps for various parts of chicken (a) and human (b) STING in the apo state, and cGAMP-bound chicken STING (c).
Extended Data Fig. 8 Data collection and model statistics.
a, Data collection and model refinement statistics. b, FSC curves between the maps and models.
Supplementary information
Supplementary Figures
Supplementary Figure 1: Original uncropped images of gels or blots.
Video 1
Morph video showing the transition from the inactive to the active state of chSTING induced by cGAMP.
Rights and permissions
About this article
Cite this article
Shang, G., Zhang, C., Chen, Z.J. et al. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP–AMP. Nature 567, 389–393 (2019). https://doi.org/10.1038/s41586-019-0998-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0998-5
This article is cited by
-
O-GlcNAc of STING mediates antiviral innate immunity
Cell Communication and Signaling (2024)
-
Universal STING mimic boosts antitumour immunity via preferential activation of tumour control signalling pathways
Nature Nanotechnology (2024)
-
Single-molecule localization microscopy reveals STING clustering at the trans-Golgi network through palmitoylation-dependent accumulation of cholesterol
Nature Communications (2024)
-
Optineurin provides a mitophagy contact site for TBK1 activation
The EMBO Journal (2024)
-
A conserved ion channel function of STING mediates noncanonical autophagy and cell death
EMBO Reports (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.