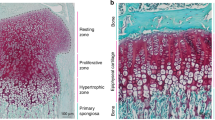
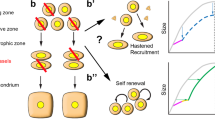
Abstract
Longitudinal bone growth in children is sustained by growth plates, narrow discs of cartilage that provide a continuous supply of chondrocytes for endochondral ossification1. However, it remains unknown how this supply is maintained throughout childhood growth. Chondroprogenitors in the resting zone are thought to be gradually consumed as they supply cells for longitudinal growth1,2, but this model has never been proved. Here, using clonal genetic tracing with multicolour reporters and functional perturbations, we demonstrate that longitudinal growth during the fetal and neonatal periods involves depletion of chondroprogenitors, whereas later in life, coinciding with the formation of the secondary ossification centre, chondroprogenitors acquire the capacity for self-renewal, resulting in the formation of large, stable monoclonal columns of chondrocytes. Simultaneously, chondroprogenitors begin to express stem cell markers and undergo symmetric cell division. Regulation of the pool of self-renewing progenitors involves the hedgehog and mammalian target of rapamycin complex 1 (mTORC1) signalling pathways. Our findings indicate that a stem cell niche develops postnatally in the epiphyseal growth plate, which provides a continuous supply of chondrocytes over a prolonged period.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The LCM-seq dataset generated during the study has been deposited at the Gene Expression Omnibus with accession number GSE113982. Numerical source data for the figures are provided in the Supplementary Information and all original confocal scans are available upon request to the authors.
References
Hall, B. K. Bones and Cartilage: Developmental and Evolutionary Skeletal Biology 2nd edn (Academic Press, Cambridge, MA, 2015).
Moss-Salentijn, L., Moss, M. L., Shinozuka, M. & Skalak, R. Morphological analysis and computer-aided, three dimensional reconstruction of chondrocytic columns in rabbit growth plates. J. Anat. 151, 157–167 (1987).
Nakamura, E., Nguyen, M. T. & Mackem, S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreERT to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 235, 2603–2612 (2006).
Snippert, H. J. et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell 143, 134–144 (2010).
Gerber, H.-P. et al. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nat. Med. 5, 623–628 (1999).
Joost, S. et al. Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 3, 221–237 (2016).
Rompolas, P. et al. Spatiotemporal coordination of stem cell commitment during epidermal homeostasis. Science 352, 1471–1474 (2016).
Suda, T., Takubo, K. & Semenza, G. L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310 (2011).
Clevers, H. The intestinal crypt, a prototype stem cell compartment. Cell 154, 274–284 (2013).
Schofield, R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells 4, 7–25 (1978).
Morrison, S. J. & Spradling, A. C. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell 132, 598–611 (2008).
Nichterwitz, S. et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat. Commun. 7, 12139 (2016).
Gilbert, S. F. Developmental Biology 11th edn (Sinauer Associates, Sunderland, MA, 2017).
Hsu, Y. C., Li, L. & Fuchs, E. Emerging interactions between skin stem cells and their niches. Nat. Med. 20, 847–856 (2014).
Li, Y. & Dudley, A. T. Noncanonical frizzled signaling regulates cell polarity of growth plate chondrocytes. Development 136, 1083–1092 (2009).
de Andrea, C. E. et al. Primary cilia organization reflects polarity in the growth plate and implies loss of polarity and mosaicism in osteochondroma. Lab. Invest. 90, 1091–1101 (2010).
Rowinsky, E. K. & Donehower, R. C. Paclitaxel (Taxol). N. Engl. J. Med. 332, 1004–1014 (1995).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Rompolas, P. et al. Live imaging of stem cell and progeny behaviour in physiological hair-follicle regeneration. Nature 487, 496–499 (2012).
Emons, J., Chagin, A. S., Sävendahl, L., Karperien, M. & Wit, J. M. Mechanisms of growth plate maturation and epiphyseal fusion. Horm. Res. Paediatr. 75, 383–391 (2011).
Kimura, H., Ng, J. M. Y. & Curran, T. Transient inhibition of the Hedgehog pathway in young mice causes permanent defects in bone structure. Cancer Cell 13, 249–260 (2008).
Everson, J. L., Fink, D. M., Chung, H. M., Sun, M. R. & Lipinski, R. J. Identification of sonic hedgehog-regulated genes and biological processes in the cranial neural crest mesenchyme by comparative transcriptomics. BMC Genomics 19, 497 (2018).
Maeda, Y. et al. Indian Hedgehog produced by postnatal chondrocytes is essential for maintaining a growth plate and trabecular bone. Proc. Natl Acad. Sci. USA 104, 6382–6387 (2007).
Newton, P. T., Xie, M., Medvedeva, E. V., Sävendahl, L. & Chagin, A. S. Activation of mTORC1 in chondrocytes does not affect proliferation or differentiation, but causes the resting zone of the growth plate to become disordered. Bone Rep. 8, 64–71 (2018).
Ahn, S. & Joyner, A. L. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell 118, 505–516 (2004).
Livet, J. et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature 450, 56–62 (2007).
Kwiatkowski, D. J. et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11, 525–534 (2002).
Sengupta, S., Peterson, T. R., Laplante, M., Oh, S. & Sabatini, D. M. mTORC1 controls fasting-induced ketogenesis and its modulation by ageing. Nature 468, 1100–1104 (2010).
Madisen, L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010).
Hall, A. P., Westwood, F. R. & Wadsworth, P. F. Review of the effects of anti-angiogenic compounds on the epiphyseal growth plate. Toxicol. Pathol. 34, 131–147 (2006).
Kaucka, M. et al. Analysis of neural crest-derived clones reveals novel aspects of facial development. Sci. Adv. 2, e1600060 (2016).
Salic, A. & Mitchison, T. J. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl Acad. Sci. USA 105, 2415–2420 (2008).
Podgorny, O., Peunova, N., Park, J. H. & Enikolopov, G. Triple S-phase labeling of dividing stem cells. Stem Cell Reports 10, 615–626 (2018).
Reibring, C.-G. et al. Expression patterns and subcellular localization of carbonic anhydrases are developmentally regulated during tooth formation. PLoS ONE 9, e96007 (2014).
Chagin, A. S. et al. G-protein stimulatory subunit alpha and Gq/11α G-proteins are both required to maintain quiescent stem-like chondrocytes. Nat. Commun. 5, 3673 (2014).
Xiao, P. et al. Sipa1 deficiency-induced bone marrow niche alterations lead to the initiation of myeloproliferative neoplasm. Blood Adv. 2, 534–548 (2018).
Nichterwitz, S., Benitez, J. A., Hoogstraaten, R., Deng, Q. & Hedlund, E. LCM-seq: a method for spatial transcriptomic profiling using laser capture microdissection coupled with polyA-based RNA sequencing. Methods Mol. Biol. 1649, 95–110 (2018).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Fan, J. et al. Characterizing transcriptional heterogeneity through pathway and gene set overdispersion analysis. Nat. Methods 13, 241–244 (2016).
Kharchenko, P. V., Silberstein, L. & Scadden, D. T. Bayesian approach to single-cell differential expression analysis. Nat. Methods 11, 740–742 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Hintze, J. L. & Nelson, R. D. Violin plots : a box plot–density trace synergism. Am. Stat. 52, 181–184 (1998).
Acknowledgements
We thank H. M. Kronenberg (MGH) for critical reading of this manuscript, O. Dobre (Blue Insight) for programming the web simulator, O. Kharchenko for graphical work (Fig. 3j), and M. E. Kastriti and O. Podgorny for methodological advice. This work was supported financially by the Swedish Research Council (A.S.C., K.F., L. Sävendahl, E.H., M.K., I.A., V.D., J.P., A.G.-L.), Karolinska Institute (A.S.C., L.L., M.K., I.A.), StratRegen KI and Stiftelsen Konung Gustaf V:s 80-årsfond (A.S.C.), Swedish Cancer Society (M.K.), the Swiss National Science Foundation (C.S.), EMBO (M.X.), Stiftelsen Frimurare Barnhuset and Sallskapet Barnavard (P.T.N.) and the Grant Agency of the Czech Republic (#14-37368G, M.H.). We also acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation, the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure.
Reviewer information
Nature thanks Ophir Klein, Michael Longaker and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
P.T.N. and A.S.C. conceived the study. P.T.N. performed the majority of the experiments with the help of L.L., B.Z., L. Sandhow, M.X., S.S., X.S., H.Q., M.H., T.B., M.E.S., A.G.-L., J.P., J.L., A.M., C.S., V.D. and E.I. A.V.A. carried out all the bioinformatics analysis. I.A., G.E., L. Sävendahl, E.H., K.F. and M.K. contributed intellectually throughout the project. P.T.N. and A.S.C. wrote the manuscript. All of the authors critically reviewed this manuscript and approved the final draft. More details are provided in the Supplementary Information.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Clonality and growth rate dynamics during maturation of the growth plate.
a–d, Clonal pattern in Col2-creERT:R26R-Confetti mice traced from P30 to P40 (a; the humerus is shown, all other images throughout the manuscript show tibial growth plates), from E14.5 to P4 (b), from P3 to P18 (c) and from P30 to P44 (d). Arrowheads in a and b show typical clones. e, Col2-creERT:R26R-Confetti mice received tamoxifen at either 3 or 30 days of age and were traced simultaneously for 4, 9 or 14 days. Clone size was quantified using Imaris. n = 3 mice per age per time point; mean ± s.e.m.; one-way ANOVA with Tukey multiple comparisons. f, Chondrocyte proliferation rate (single EdU injection, 24 h before analysis) was quantified at different ages. Data are mean ± s.e.m.; n = 3 mice per age. g, The rate of bone formation under the growth plate of proximal tibia or distal femur was assessed by xylenol pulse-chase bone labelling. n = 2–4 mice per time point; mean ± s.e.m. h, Representative histological images (haemotoxylin and eosin staining) of the proximal tibial growth plate before (P3, left), during (P10, middle) and after (P30, right) SOC formation. Blue dashed lines demarcate borders between the resting, proliferating and hypertrophic zones. The junction between SOC and the resting zone is magnified in inserts. i, Col2-creERT:R26R-Confetti mice traced from P30 to 6 months of age. Arrowheads indicate stable clones and the arrow indicates a labelled resting-zone cell, which did not form a column during the tracing period. j, Col2-creERT:R26R-Confetti mice traced from E14.5 to P70. The black or white dashed lines demarcate the growth plate from the surrounding tissues. Data in a–d and j represent two independent experiments or litters, data in e–h represent one experiment per time point (with the number of mice per data point indicated above), whereas data in i represent 3 independent experiments or litters.
Extended Data Fig. 2 Contribution of slowly dividing resting-zone cells to the underlying growth-plate zones.
a, Mice were injected with EdU on P3 (two doses), CldU on P10 and IdU on P12, 4 h before tissue collection. Confocal images of stained sections are presented, with individual channels shown in the top row, and channels merged to the right. The merged image is labelled with arrowheads indicating double positive cells (E, EdU; I, IdU; C, CldU) and area in a′ expanded below to show a close-up image of double- and triple-positive cells (b′–d′). The second row in a shows domains containing each label and the overlapping areas. b, LRCs were identified in H2B-GFP mice (Tg(tetO-HIST1H2BJ/GFP)47Efu/J mice crossed with B6.Cg-Gt(ROSA)26Sortm1(rtTA∗M2)Jae/J), exposed to doxycycline from E14.5 to P2, and chased until P2, P9, P18 or P28. LRCs appear green, whereas nuclei were visualized with propidium iodide (red). Arrowheads indicate typical GFP-retaining cells in the resting zone. Laser power was enhanced for older mice in order to obtain comparable GFP intensity in confocal scans. c, d, H2B-GFP mice were crossed with Col2-creERT:R26R-Confetti mice and column-forming LRCs were visualized with doxycycline being administered from E17.5 ± 1 day until P3 ± 1 and tamoxifen on P5. Colocalization of GFP-retaining cells with Confetti clones (arrowheads) were quantified at P21 (c) and P28 (d). Dashed lines in b–d demarcate the border between cartilage and the surrounding tissues. The SOC, resting (R), proliferating (P), and hypertrophic (H) zones are labelled for orientation. Data in a represents two independent experiments, data in c and d represent one experiment per time point.
Extended Data Fig. 3 Changes in the expression profile of the resting zone upon niche formation.
a, Sections of cartilage at P2 and P28 were imaged before and after cells were captured by LCM for subsequent RNA-seq analysis. b, c, Gene ontology categories enriched by genes significantly upregulated (b) or downregulated (c) in the resting zone at P28 compared to P2. Logarithmic P values of significance are indicated on the x axis. d, Expression heat map for the genes most significantly up- and downregulated between P2 and P28 time points in the resting zone (DESeq2 test with multiple testing correction, FDR < 0.005). e, Heat map of similarities in gene expression between individual samples from the resting and hypertrophic zones from P2 and P28 time points. For each sample, expression of non-redundant PAGODA aspects was calculated and similarity between samples was defined as correlation of aspect expressions (see Methods for details). f–h, Proximal tibial growth plates from P2, P9, P18 and P28 mice were immunostained for CD73 (f), Ki67 (g) or MEF2C (h) and counterstained with DAPI. For a–e, a total of 8 pools of cells from 7 mice were collected at P2 and 7 pools of cells from 7 mice were collected at P28. For f–h, staining was performed in a single experiment (minimum 3 mice) per time point.
Extended Data Fig. 4 Isolation and ex vivo differentiation of CD49e+CD73+ resting-zone stem cells.
a, Isolated growth-plate chondrocytes were analysed by FACS and the live cells were sorted for expression of both CD73 and CD49e. FMO control for CD49e (a′), FMO control for CD73 antibody (a′′) and gating strategy for both CD49e and CD73 (a′′′); percentage of single (CD49e+CD73−) and double (CD49e+CD73+) positive cells among all live cells. b, c, The CD49e+CD73+ cells were grown in chondrogenic conditions in pellets ex vivo, and thereafter stained either with safranin-O (b) or for type-II collagen by immunofluorescence (c). d–f, Alizarin red staining (d) indicated that CD49e+CD73+ cells cultured in osteoblastic-differentiation medium differentiated to osteoblasts similarly to mesenchymal stem cells (MSCs) (e), but not in the absence of differentiation medium (f). g–i, Oil red O staining indicated that CD49e+CD73+ cells cultured in adipogenic-differentiation medium differentiated to adipocytes (g), albeit to a lesser extent than MSCs (h), but not in the absence of differentiation medium (i). Data represents two independent experiments.
Extended Data Fig. 5 Orientation of cell divisions in the epiphyseal niche.
a, Col2-creERT:R26R-Confetti mice were traced as indicated above each plot. The angle between cells in self-standing dyads within the resting zone was measured relative to the longitudinal axis of the bone (2–5 mice per tracing period, with a total of 559 dyads analysed). b, H2B-GFP mice were labelled with doxycycline from E14.5 to P2 and chased until P28 to visualize slow-dividing cells (green). Cilia were visualized by immunostaining for acetylated tubulin (arrowheads). c, Col2-creERT:R26R-Confetti mice were traced from P3 to P33 and immunostained for PAR3. Arrowhead indicates a PAR3-positive clonal dyad. d, Growth plates from 28-day-old mice were immunostained for PAR3. e, H2B-GFP mice chased as in b were immunostained for PKC-ζ. Arrowheads indicate positively stained cells in the resting zone. f, g, To block spindle degradation, mice were treated with Taxol (10 mg kg−1) for two days before tissue collection at P30 and subsequently immunostained for PAR3 (f) or PKC-ζ (g). Arrowheads in f highlight even (left panel) and uneven (right panel) distribution of PAR3. Even with Taxol treatment, staining for PKC-ζ in the resting zone was minimal (arrowheads indicate positive cells). h, Growth plates from 28-day-old H2B-GFP mice chased as in b were stained for phosphorylated histone H3 (p-H3). i, Blunted columns (arrowheads) in Col2-creERT:R26R-Confetti mice traced from P30 to P45. The dashed lines demarcate the growth plate from surrounding tissues. The resting (R), proliferating (P), hypertrophic (H) zones and SOC are indicated for orientation. Data in b, e–h each represent one experiment (minimum 3 mice per experiment); data in c, d and i represent two independent experiments.
Extended Data Fig. 6 Indications that hedgehog signalling controls stem cell renewal in the epiphyseal niche.
a, Shh-GFP reporter mice revealed Shh expression within the SOC at P30 (blue staining, DAPI). b, In situ hybridization for Gli2 mRNA at P30 (arrowheads point towards typical labelling). c, Gli1-CreERT2:tdTomato mice traced from P29 to P33 to reveal Gli1 activity. d, Col2-creERT:R26R-Confetti mice were traced at P30–P35, and received six injections of either DMSO or the SMO antagonist vismodegib (Vism, 100 mg per kg(body weight) per injection) between P31 and P34. Blue dashed lines indicate the top 50 µm, where the clone size was quantified (d′′′, mean ± s.e.m., n = 123 clones from 8 mice (vismodegib); n = 49 from 4 mice (DMSO)). When vismodegib treatment was extended by two extra injections, the growth plate fused at P37 (d′′′′; haemotoxylin and eosin staining). e–h, Growth plates from mice treated as in d′ and d′′ were analysed for proliferation within the top 50 µm (demarcated by blue dashed line) by means of EdU incorporation (e; injected daily P32–P34; n = 4 for each treatment) and KI67 immunolabelling (f; n = 4 (DMSO) or 3 (Vism) mice) and for expression of differentiation marker MEF2C (g) and stem cell marker CD73 (h). Arrowheads in e–g indicate typical positively-stained cells. i, Mice received six doses of either DMSO or SMO agonist (SAG, 25 mg per kg(body weight) per injection) between P31 and P34 and proliferation was analysed by Ki67 immunolabelling at P35 and quantified in the top 50 µm (depicted by blue dashed lines, n = 4 (DMSO) or 3 (SAG) mice). j, Mice at 7 ± 1 weeks of age received a single intra-articular administration of either SAG or DMSO followed by intraperitoneal EdU injection 4 h later. EdU incorporation was quantified 20 h later in the top 50 µm of the growth plate (n = 6). k, CD73 was immunodetected in mice treated as in i. CD73+CD49e+ cells were sorted by FACS as in Extended Data Fig. 4a and colony size (l) and number (m) were assessed following 10 days of culture in the presence of vehicle (DMSO (or ethanol for GANT61)), recombinant mouse sonic hedgehog (rmShh, 1 µg ml−1), vismodegib (1 µM) or the Gli1 inhibitor GANT61 (5 µM). Data in l and m are mean ± s.e.m., n = 9 wells from 3 independent experiments for rmShh and Vism, and n = 3 wells from one experiment (GANT61). All white dashed lines demarcate the growth plate. The SOC, the resting (R), proliferating (P) and hypertrophic (H) zones are indicated for orientation. Data in a and j represent two independent experiments, data in b, c, d′′′′, e and k each derive from one experiment and those in d′–d′′′ are pooled from three independent experiments with the number of mice in each experiment indicated above. Plots represent mean ± s.e.m. and were analysed by unpaired two-tailed t-test, except the comparison between DMSO, rmShh and Vism groups (l and m), which was by one-way ANOVA with Tukey’s multiple comparisons.
Extended Data Fig. 7 Identification of Shh-positive cell populations within the bone marrow.
a, Representative FACS profile showing gating strategy for analysing expression of Shh–GFP+ cells in bone marrow. The cells were gated within propidium iodide-negative live cells. b, Distribution of GFP+ cells in central marrow and endosteal region. The cells from central marrow were obtained by gently crushing bones and the endosteal cells were obtained by enzyme dissociation of bone fragments. c, Percentages of CD41+ megakaryocytes and haematopoietic progenitor (KIT+) cells among CD45+GFP+ cells. d, Percentages of GFP+ cells within haematopoietic cells (CD45+), endothelial cells (CD31+), mesenchymal progenitor cells (MPC, CD45−CD31−SCA1−CD51+) and MSCs (CD45−CD31−SCA1+CD51+). e, Representative FACS profile showing GFP+ cells among total live cells. In b–d, each data point represents one mouse, and the mean is shown as a line. Two independent analysis were performed for b–e with at least 3 GFP+ mice for each experiment.
Extended Data Fig. 8 Postnatal cartilage-specific Tsc1 ablation does not promote hypertrophic differentiation in the resting zone.
All Col2-creERT:R26R-Confetti:Tsc1 mice were injected with tamoxifen on P3 and analysed either on P28 (a, b, h) or P40 (c–g, i). The central sections of the tibial growth plates of Col2-creERT:R26R-Confetti:Tsc1fl/+ (Tsc1+/−, left) and Col2-creERT:R26R-Confetti:Tsc1fl/fl (Tsc1−/−, right) mice were imaged on P28 (a) and the clonal size of round cells above the columnar zone quantified with Imaris (b; mean ± s.e.m.; n = 3 (Tsc1−/+) and 6 (Tsc1−/−), unpaired two-tailed t-test). c, d, Haemotoxylin and eosin staining of the sections depicted in Fig. 3a, b. Arrowheads indicate typical clusters in the resting zone of Tsc1−/− mice. e–g, In situ hybridization for ColX (e) and Ihh (f), and immunostaining for Shh/Ihh (g). Insets in e and f show a magnified view of the outlined area in the resting zone, showing absence of staining. h, In situ hybridization for Gli2. i, Immunostaining for phosphorylated S6 (readout for mTORC1 activity). Typical positive cells in g–i are indicated by arrowheads. The dashed lines demarcate the growth plate. The SOC, the resting (R), proliferating (P), and hypertrophic (H) zones are labelled for orientation. Data in a, b and i derive from 2 independent experiments, whereas c–h represent one experiment with several mice stained at least twice.
Extended Data Fig. 9 Postnatal cartilage-specific Tcs1 ablation does not elevate proliferation in the resting zone.
One (Tsc1+/–) or two (Tsc1−/−) copies of the Tsc1 gene were deleted in Col2-creERT:R26R-Confetti:Tsc1-floxed mice by tamoxifen injection on P3, and mice were analysed on P40. a–c, EdU was injected once daily for four days before tissue collection, and EdU-positive cells in the resting (c, left), proliferating (c, middle) and hypertrophic (c, right) zones were quantified (mean ± s.e.m.; n = 3 (Tsc1+/−) and 7 or 8 (Tsc1−/−)). d, e, Immunostaining for Ki67 (d) and phospho-histone H3 markers of proliferation (e). f, Mice were injected with a single dose of EdU 4 h before tissue collection, and the percentage of EdU-positive cells in the proliferating zone was calculated (mean ± s.e.m.; n = 3). g, Immunostaining for Sox9 (enlarged images on the right of each pair are shown without the DAPI channel, to improve visualization). h, Immunostaining for acetylated tubulin (the panels to the right are close up images of a single plane, to aid visualization of the cilia), with the mean angle of cilia in the top 100 µm indicated in the graph (mean ± s.e.m.; n = 3 (Tsc1+/−) and 4 (Tsc1−/−)). Counterstaining with DAPI (blue) and two-tailed unpaired t-test were used throughout experiments presented. The SOC, resting (R), proliferating (P) and hypertrophic (H) zones are labelled for orientation. The white dashed lines demarcate the growth plate. The blue dashed lines in h indicate the top 100 µm of the growth plate. Data from a–e and h each derive from 2 independent experiments, and f and g are from a single experiment each with n = 3 mice.
Extended Data Fig. 10 mTORC1 signalling controls symmetric versus asymmetric division of resting zone cells.
a, Immunostaining for PAR3 combined with confocal visualization of Confetti clones in the resting zone of Col2-creERT:R26R-Confetti:Tsc1fl/+ (Tsc1+/−) or Col2-creERT:R26R-Confetti:Tsc1fl/fl (Tsc1−/−) mice injected with tamoxifen at P3 was performed at P40. Representative images for asymmetric and symmetric distribution of PAR3 within clones are shown (arrowheads) and distribution within the resting zone clones was quantified (mean ± s.e.m., n = 4 mice; unpaired two-tailed t-test). Dashed lines demarcate PAR3–Confetti double-positive dyads. b, Tsc1 was ablated by tamoxifen injection on P3, and from P40, mice were given either vehicle (DMSO) or the SMO antagonist vismodegib (8 doses, twice daily) before analysis at P45. All mice were injected daily with axitinib to prevent growth plate fusion. The growth plates were imaged for Confetti proteins (b′), or immunostained for CD73 (b′′′, arrowheads indicate CD73-positive cells within the proliferative zone) and Ki67 (b′′′′, arrowheads indicate typical Ki67-positive cells). b′′, The width of Confetti-labelled clones at 100 µm from the SOC surface was quantified (mean ± s.e.m.; Tsc1+/− with DMSO (n = 3 mice), Tsc1−/− with DMSO (n = 4 mice), Tsc1−/− with Vism (n = 4 mice); one-way ANOVA with Tukey’s multiple comparisons). The data derives from 1 (Tsc1+/− group) or 3 (Tsc1−/− groups) independent experiments. c, Col2-creERT:R26R-Confetti:Raptorfl/+ (Raptor+/−) or Col2-creERT:R26R-Confetti:Raptorfl/fl (Raptor−/−) mice received tamoxifen on P3 and were analysed on P40. The clonal pattern revealed blunted columns (arrowheads) and a decreased number of labelled clones within the growth plate of Raptor−/− mice as counted at the horizontal midline (c′′′; mean ± s.e.m., n = 3 mice from 1 (Raptor+/−) or 2 (Raptor−/−) independent litters; unpaired two-tailed t-test). The dashed lines in b and c demarcate growth plates. The SOC, the resting (R), proliferating (P), and hypertrophic (H) zones are labelled for orientation.
Supplementary information
Supplementary Information
This file contains the web-simulator instructions which provide detailed instructions explaining how to model cellular dynamics in the epiphyseal growth plate, Supplementary Tables 1–5 and a detailed Author Contributions statement.
Video 1: Clonal tracing of chondrocytes prior SOC maturation revealed small clones.
Col2CreERt:R26-Confetti mice were labelled at embryonic day 14.5 and then traced for 10 days until P4. The video represents 2 independent experiments.
Video 2: Clonal tracing of chondrocytes after SOC maturation revealed large clones.
Col2CreERt:R26-Confetti mice were labelled at P30 and then traced for 10 days until P40. The video represents 2 independent experiments.
Video 3: Clonal clusters of chondro-progenitors appear in the resting zone upon Tsc1 ablation.
Col2CreERt:R26-Confetti:Tsc1-fl/fl mice received tamoxifen on P3 and from P40 were given vehicle (DMSO, 8 doses, twice each day) prior to analysis at P45. Mice received daily axitinib to prevent fusion of the growth plate. The video represents 3 independent experiments.
Video 4: Clonal clusters of chondro-progenitors in Tsc1 cKO mice form columns upon hedgehog inhibition.
Col2CreERt:R26-Confetti:Tsc1-fl/fl mice received tamoxifen on P3 and from P40 were given SMO antagonist (vismodegib, 8 doses, twice each day) prior to analysis at P45. Mice received daily axitinib to prevent fusion of the growth plate. The video represents 3 independent experiments.
Source data
Rights and permissions
About this article
Cite this article
Newton, P.T., Li, L., Zhou, B. et al. A radical switch in clonality reveals a stem cell niche in the epiphyseal growth plate. Nature 567, 234–238 (2019). https://doi.org/10.1038/s41586-019-0989-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0989-6
This article is cited by
-
Compensatory growth and recovery of cartilage cytoarchitecture after transient cell death in fetal mouse limbs
Nature Communications (2024)
-
The level of protein in the maternal murine diet modulates the facial appearance of the offspring via mTORC1 signaling
Nature Communications (2024)
-
Gli1 labels progenitors during chondrogenesis in postnatal mice
EMBO Reports (2024)
-
The NAD salvage pathway in mesenchymal cells is indispensable for skeletal development in mice
Nature Communications (2023)
-
Premature aging of skeletal stem/progenitor cells rather than osteoblasts causes bone loss with decreased mechanosensation
Bone Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.