Abstract
The cytokine interferon-γ (IFNγ) is a central coordinator of innate and adaptive immunity, but its highly pleiotropic actions have diminished its prospects for use as an immunotherapeutic agent. Here, we took a structure-based approach to decoupling IFNγ pleiotropy. We engineered an affinity-enhanced variant of the ligand-binding chain of the IFNγ receptor IFNγR1, which enabled us to determine the crystal structure of the complete hexameric (2:2:2) IFNγ–IFNγR1–IFNγR2 signalling complex at 3.25 Å resolution. The structure reveals the mechanism underlying deficits in IFNγ responsiveness in mycobacterial disease syndrome resulting from a T168N mutation in IFNγR2, which impairs assembly of the full signalling complex. The topology of the hexameric complex offers a blueprint for engineering IFNγ variants to tune IFNγ receptor signalling output. Unexpectedly, we found that several partial IFNγ agonists exhibited biased gene-expression profiles. These biased agonists retained the ability to induce upregulation of major histocompatibility complex class I antigen expression, but exhibited impaired induction of programmed death-ligand 1 expression in a wide range of human cancer cell lines, offering a route to decoupling immunostimulatory and immunosuppressive functions of IFNγ for therapeutic applications.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Structure factors and coordinates have been deposited in the Protein Data Bank with identification numbers 6E3K and 6E3L. Diffraction images have been deposited in the SBGrid Data Bank with dataset ID 591 and 592. Next-generation sequencing data files from the human transcriptome study were deposited to the NCBI Gene Expression Omnibus (GEO) data repository with accession number GSE122672. Other data and materials are available upon request from the corresponding author.
References
Pace, J. L., Russell, S. W., LeBlanc, P. A. & Murasko, D. M. Comparative effects of various classes of mouse interferons on macrophage activation for tumor cell killing. J. Immunol. 134, 977–981 (1985).
Nakajima, C. et al. A role of interferon-γ (IFN-γ) in tumor immunity: T cells with the capacity to reject tumor cells are generated but fail to migrate to tumor sites in IFN-γ-deficient mice. Cancer Res. 61, 3399–3405 (2001).
Stark, G. R., Kerr, I. M., Williams, B. R., Silverman, R. H. & Schreiber, R. D. How cells respond to interferons. Annu. Rev. Biochem. 67, 227–264 (1998).
Mandai, M. et al. Dual faces of IFNγ in cancer progression: a role of PD-L1 induction in the determination of pro- and antitumor immunity. Clin. Cancer Res. 22, 2329–2334 (2016).
Yphantis, D. A. & Arakawa, T. Sedimentation equilibrium measurements of recombinant DNA derived human interferon gamma. Biochemistry 26, 5422–5427 (1987).
Bach, E. A. et al. Ligand-induced autoregulation of IFN-γ receptor β chain expression in T helper cell subsets. Science 270, 1215–1218 (1995).
Pernis, A. et al. Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science 269, 245–247 (1995).
Tau, G. Z., Cowan, S. N., Weisburg, J., Braunstein, N. S. & Rothman, P. B. Regulation of IFN-γ signaling is essential for the cytotoxic activity of CD8+ T cells. J. Immunol. 167, 5574–5582 (2001).
Walter, M. R. et al. Crystal structure of a complex between interferon-γ and its soluble high-affinity receptor. Nature 376, 230–235 (1995).
Blouin, C. M. et al. Glycosylation-dependent IFN-γR partitioning in lipid and actin nanodomains is critical for JAK activation. Cell 166, 920–934 (2016).
Krause, C. D. et al. Seeing the light: preassembly and ligand-induced changes of the interferon γ receptor complex in cells. Mol. Cell. Proteomics 1, 805–815 (2002).
Moraga, I. et al. Tuning cytokine receptor signaling by re-orienting dimer geometry with surrogate ligands. Cell 160, 1196–1208 (2015).
Roder, F., Wilmes, S., Richter, C. P. & Piehler, J. Rapid transfer of transmembrane proteins for single molecule dimerization assays in polymer-supported membranes. ACS Chem. Biol. 9, 2479–2484 (2014).
Richter, D. et al. Ligand-induced type II interleukin-4 receptor dimers are sustained by rapid re-association within plasma membrane microcompartments. Nat. Commun. 8, 15976 (2017).
Mendoza, J. L. et al. The IFN-λ–IFN-λR1–IL-10Rβ complex reveals structural features underlying type III IFN functional plasticity. Immunity 46, 379–392 (2017).
Thiel, D. J. et al. Observation of an unexpected third receptor molecule in the crystal structure of human interferon-γ receptor complex. Structure 8, 927–936 (2000).
Mikulecký, P. et al. Crystal structure of human interferon-γ receptor 2 reveals the structural basis for receptor specificity. Acta Crystallogr. D 72, 1017–1025 (2016).
Vogt, G. et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 37, 692–700 (2005).
Lundell, D., Lunn, C. A., Senior, M. M., Zavodny, P. J. & Narula, S. K. Importance of the loop connecting A and B helices of human interferon-γ in recognition by interferon-γ receptor. J. Biol. Chem. 269, 16159–16162 (1994).
Lunn, C. A. et al. A point mutation of human interferon gamma abolishes receptor recognition. Protein Eng. 5, 253–257 (1992).
Thomas, C. et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell 146, 621–632 (2011).
Levin, A. M. et al. Exploiting a natural conformational switch to engineer an interleukin-2 ‘superkine’. Nature 484, 529–533 (2012).
Brideau-Andersen, A. D. et al. Directed evolution of gene-shuffled IFN-α molecules with activity profiles tailored for treatment of chronic viral diseases. Proc. Natl Acad. Sci. USA 104, 8269–8274 (2007).
Bandaranayake, A. D. et al. Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors. Nucleic Acids Res. 39, e143 (2011).
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D 68, 352–367 (2012).
Smart, O. S. et al. Exploiting structure similarity in refinement: automated NCS and target-structure restraints in BUSTER. Acta Crystallogr. D 68, 368–380 (2012).
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006).
Karplus, P. A. & Diederichs, K. Assessing and maximizing data quality in macromolecular crystallography. Curr. Opin. Struct. Biol. 34, 60–68 (2015).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D 66, 12–21 (2010).
Wilmes, S. et al. Receptor dimerization dynamics as a regulatory valve for plasticity of type I interferon signaling. J. Cell Biol. 209, 579–593 (2015).
Ho, C. C. M. et al. Decoupling the functional pleiotropy of stem cell factor by tuning c-Kit signaling. Cell 168,1041–1052 (2015).
Serge, A., Bertaux, N., Rigneault, H. & Marguet, D. Dynamic multiple-target tracing to probe spatiotemporal cartography of cell membranes. Nat Methods 5, 687–694 (2008).
Moraga, I. et al. Synthekines are surrogate cytokine and growth factor agonists that compel signaling through non-natural receptor dimers. eLife 6, e22882 (2017).
Bern, M., Kil, Y. J. & Becker, C. Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics 40, 13.20.1–13.20.14 (2012).
Acknowledgements
We thank W. Schneider, H.-H. Hoffman and C. Rice for assistance with antiviral experiments; S. Bendall and L. Borges for assistance with CyTOF experiments; and J.-L. Casanova, J. Bustamante and C. Oleaga for assistance with experiments with IFNGR2 T168N cell lines.This work was supported by NIH grants 1U19AI109662, 5R01CA177684 and NIH RO1-AI51321 (to K.C.G.), by the DFG grants SFB 944 and PI 405/10-1 (to J.P.), by NIH HD090156 (to R.S.H.), and by NIH U54 CA209971 and DoD BC140436 (to E.G.E.). K.C.G. is an investigator of the Howard Hughes Medical Institute and is supported by the Ludwig Institute and the Younger Family Chair. J.L.M. is supported by NIH award K01CA175127. We thank the staff at Stanford Synchrotron Radiation Lightsource and Advanced Light Source for their assistance. The Advanced Light Source is a Department of Energy Office of Science User Facility under Contract No. DE-AC02-05CH11231. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393).
Reviewer information
Nature thanks Michael Parker, Antoni Ribas and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Author information
Authors and Affiliations
Contributions
J.L.M. and K.C.G. conceived and designed the project and wrote the manuscript. J.L.M. and T.M.H. designed and performed preliminary yeast display experiments. J.L.M. engineered IFNγR1, designed and prepared all recombinant proteins for the study, crystallized the complexes, solved the structure, and carried out SPR and cytokine secretion assay. J.L.M. and K.M.J. refined the structures. J.L.M. engineered signalling ligands and measured affinities by SPR. N.T. cloned and expressed receptors for SPR and helped J.L.M. with SEC experiments characterizing receptor binding properties of the GIFNs. J.S.B. performed single-molecule TIRF measurements. J.L.M. and L.S. performed cytokine secretion assays and screened cancer cell lines for MHC I:PD-L1 bias. N.K.E. performed PD-L1 and MHC class I upregulation assays on A549 cells, monocytes, macrophages and dendritic cells by FACS. J.L.M. measured gene expression by qPCR and prepared samples for next-generation sequencing of a human gene expression AmpliSeq panel. S.J.B. performed mass spectrometry analysis of IFNγR2(T168N) neo N-glycosylation site. R.S.H., J.P., E.G.E. and K.C.G. supervised the research. N.K.E., K.M.J. and J.S.B. contributed equally to the studies.
Corresponding author
Ethics declarations
Competing interests
K.C.G. and J.L.M. are co-inventors on provisional patent application 62/712,128, which includes discoveries described in this manuscript. K.C.G. is the founder of Synthekine Therapeutics.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Characterization of IFNγ complex formation and stabilizing mutations.
a, Schematic for quantifying the homodimerization of either IFNγR1 (top) or IFNγR2 (bottom) using dye-labelled anti-GFP nanobodies labelled with Rho11 and DY647. b, Homodimerization of IFNγR1 in the absence and presence of ligand. Data are mean ± s.e.m.; n = 8 (−IFNγ) and 12 (+IFNγ); n refers to biologically independent samples. c, Homodimerization of IFNγR2 in absence and presence of ligand. Data are mean ± s.e.m.; where n = 5 (−IFNγ) and 16 (+IFNγ); n refer to biologically independent samples. d, IFNγR1 displays on the surface of the yeast. Second from left, anti-Myc-647 antibody; far left, steptavidin–Alexa Fluor 647. The high avidity form of IFNγR2 only binds to IFNγR1 in the presence of IFNγ (far right) and does not bind IFNγR1 alone (second from right). Data are representative of at least 3 biologically independent experiments. e, Sequence alignment of IFNγR1 genes including 13 first-generation variants and the shuffled IFNγR1 F05 variant relative to wild-type. IFNγR1 F05 combines six mutations including Q167K and M161K. The combination of Q167K and M161K is not seen in any single first-generation mutant.
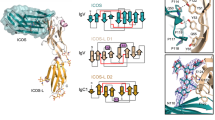
Extended Data Fig. 2 Purification and electron density maps of the IFNγ hexameric signalling complex.
a, SEC (Superdex S200 column) of the 2:2:2 IFNγ–IFNγR1–IFNγR2 complex and SDS–PAGE gels of the deglycosylated (top, left gel) and fully glycosylated (top, right gel) forms. Data shown are representative of at least 3 biologically independent experiments. mAU, milli absorbance units. b, Electron density maps showing interactions at site 2 (top) and site 3 (bottom) in the deglycosylated complex. For each pair of site 2 or site 3 panels, the left panel shows a simulated annealing composite omit map (grey) contoured at 1σ, and the right panel shows a 2mFo − DFc map (blue) calculated using phases from the final refined model and contoured at 1σ. IFNγ (green) engages IFNγR2 (cyan) at site 2, whereas the stems of IFNγR1 F05 (yellow) and IFNγR2 interact at site 3.
Extended Data Fig. 3 Quantification of IFNγR2(T168N) glycoforms by mass spectroscopy.
a, The mutant IFNγR2(T168N) protein was expressed in HEK293S GnTI− cells and purified by SEC. The SEC profile is shown (left) with the corresponding fractions on SDS–PAGE (right). Lane 1 shows the sample loaded on the SEC column, lane 2 shows the Mark 12 protein ladder, and lanes 3–9 are fractions 14–20. Data are representative of at least 3 biologically independent experiments. b, The protein coverage map shows sequence coverage of 76.82% for the entire IFNγR2(T168N) protein including the peptide of interest, containing N168, which is underlined. This peptide was detected as a glycopeptide with several glycoforms as quantified in Fig. 3d. Mappings highlighted in green indicate high confidence with a false discovery rate (FDR) below 1% and yellow indicates a FDR of 1–5%. Carbamidomethyl (C) and glycosylation (G) sites are indicated above the site of modification. Confidence levels were determined as previously described38. c, MS2 spectra confirming that the ion used for the EICs shown in Fig. 3d is the peptide SSPFDIADNSTAF from IFNγR2(T168N) modified with a HexNAc2Hex5 glycan attached to N168. The data shown are for a single experiment.
Extended Data Fig. 4 Disrupting IFNγR2 binding and characterization of IFNγ partial agonists.
a, The structure of IFNγ (blue and tan cartoon) binding site for IFNγR2 (interacting loops are shown in green). Based on the hexameric complex, positions in IFNγ at the IFNγR2 binding interface were identified to be important for binding to IFNγR2. The location of IFNγ mutations K74A, E75Y, and N83R are shown as sticks coloured in red. b, When complexed with IFNγ R1, the IFNγ triple mutant (K74A/E75Y/N83R) results in the loss of detectable binding to IFNγR2 (up to 100 μM) as determined by SPR. The titration data are from a single experiment. c, Relative co-tracking of binding of IFNγR1 (left panel) and IFNγR2 (right panel) for wild-type IFNγ and variants. GIFN2, one IFNγR1 and one IFNγR2 binding in cis; GIFN3, two IFNγR1 molecules bound; GIFN4, reduced affinity for both IFNγR1 and IFNγR2 (see Extended Data Fig. 5). Data are mean ± s.e.m.; n is indicated over each bar; n refers to the number of biologically independent samples. d, STAT1 activation of GIFN4 in Hap1 cells. Curve was fit to a first-order logistic model. Data are mean; n = 2 biologically independent samples. e, Quantification of MHC class I expression by qPCR using primers against HLA chain b in A549 treated cells. Data are mean ± s.e.m.; n = 3 biologically independent experiments. f, Dendritic cells purified from whole blood were treated for 48 h with either wild-type IFNγ or the partial agonists for 48 h; these agonists upregulate MHC class I antigen expression as quantified by fluorescently labelled antibody (left) or PD-L1 (right). Data shown are for ligand concentrations of 0.1, 0.5, 2.5, 12.5 and 62.5 nM (left to right, respectively, for each agonist). Data are mean ± s.e.m.; n = 3 biologically independent experiments. g, Using the MHC class I and PD-L1 expression data (f and Fig. 5b), the ratio of MHC I:PD-L1 induction was determined for each protein concentration relative to wild-type. Left, dendritic cells; right, A549 cells. Data are mean ± s.e.m.; n = 3 biologically independent experiments. h, Biased MHC I:PD-L1 expression for monocytes (right) and macrophages (left) isolated from PBMCs. Data as shown are for protein concentrations of 0.1, 0.5, 2.5, 12.5 and 62.5 nM (left to right, respectively, for each agonist). Data are mean ± s.e.m.; n = 6 independent samples. i, Cytokine secretion profile of IFNγ and partial agonists for PBMCs treated with 100 nM of each protein for 24 h. Shown are the mean expression of 36 secreted cytokines that are significantly different (P < 0.05) between the wild-type and PFA-treated control. Expression of IL-10, IL-12P70, IL-2, IP-10, MIG and IL-23 are indicated in text or asterisks aligned below the text. Data shown are for n = 2 biologically independent samples, each assayed in triplicate. P values were determined using the Student’s t-test with a two-tailed distribution of a two-sample heteroscedastic test.
Extended Data Fig. 5 Design and biochemical characterization of GIFNs.
a, Diagram showing the strategy for expression and purification of heterodimeric IFNγ variants. The asymmetric variants containing three tags were expressed as single polypeptides in Hi5 insect cells. The proteins were first harvested from the secreted medium with a C-terminal 8× His tag. Proteins eluted from nickel resin were then treated with 1:100 (by mass) human rhinovirus 3C protease for 24 h at 4 °C to cleave the 3C protease tag. The 3C tag is flanked by Gly-Gly-Gly-Ser motifs at both ends (G3S-3C-G3S) to ensure accessibility of the protease site between the two chains of IFNγ. The cleaved proteins were then purified using the N-terminal protein C-tag and a final 8× His tag purification to ensure isolation of the correctly paired heterodimeric IFNγ proteins. b, Table of mutations for each of the GIFN proteins, indicating the affected receptor binding sites. GIFN2, one IFNγR1 and one IFNγR2 binding in cis; GIFN3, two IFNγR1 molecules bound; GIFN4, reduced affinity for both IFNγR1 and IFNγR2. c, SEC profiles and SDS–PAGE gel fractions for 200 μg wild-type IFNγ (black) or equal molar quantities of GIFN1 (purple), GIFN2 (red), GIFN4 (orange), IFNγR1 F05 (grey), and IFNγR2 (green). Individual proteins have been purified and analysed by SEC at least three times. d, e, To determine the receptor-binding properties of the GIFN proteins, the shifts in the SEC profiles and gels were compared relative to the wild-type protein as described for c except IFNs were mixed with equimolar quantities of IFNγR1 F05 (d) or equimolar quantities of both IFNγR1 F05 and IFNγR2 (e). These data are from single experiments except the wild-type experiments which were performed at least three times.
Supplementary information
Rights and permissions
About this article
Cite this article
Mendoza, J.L., Escalante, N.K., Jude, K.M. et al. Structure of the IFNγ receptor complex guides design of biased agonists. Nature 567, 56–60 (2019). https://doi.org/10.1038/s41586-019-0988-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-019-0988-7
This article is cited by
-
Recombinant IFN-γ1b Treatment in a Patient with Inherited IFN-γ Deficiency
Journal of Clinical Immunology (2024)
-
Bio-guided isolation of potential anti-inflammatory constituents of some endophytes isolated from the leaves of ground cherry (Physalis pruinosa L.) via ex-vivo and in-silico studies
BMC Complementary Medicine and Therapies (2023)
-
Strategies to therapeutically modulate cytokine action
Nature Reviews Drug Discovery (2023)
-
LncRNA TINCR impairs the efficacy of immunotherapy against breast cancer by recruiting DNMT1 and downregulating MiR-199a-5p via the STAT1–TINCR-USP20-PD-L1 axis
Cell Death & Disease (2023)
-
Engineering cytokine therapeutics
Nature Reviews Bioengineering (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.